This is an AI for Health MSc project. Students are elgible to receive a monthly reimbursement of €500,- for a period of six months. For more information please read the requirements.
Clinical Problem
There are about 6,000 different rare, genetic disorders. For the majority of patients living with these disorders, there are currently no treatment options. Developing drugs for each of these disorders will be a tedious and costly process. Drug repurposing, the reuse of approved drugs for new indications, is an interesting treatment strategy. The clinical development trajectory is usually much shorter than for development of entirely new drugs, and the expenses for their development are lower. Deep learning may be used to predict drug repurposing candidates.
To identify more of these drug repurposing candidates, we need to make optimal use of the existing knowledge of the clinical overlap between diseases, the molecular targets of FDA-approved drugs and the genes / proteins relevant for the disease. This knowledge can be represented by a drug repurposing knowledge graph. Advanced deep learning methods that use graphs as input (graph neural networks) can be exploited to learn about new relationships because they are able to reason over the existing knowledge about drugs, their current indications and their targets. The ultimate goal of this project is to develop a framework for prioritizing drug repurposing candidates for further preclinical and clinical development. We are particularly interested in identifying drug repurposing candidates for myotonic dystrophy (DM1).
DM1 is a rare, inherited, progressive and multisystemic disorder. It is the most common adult form of muscular dystrophy, with an estimated prevalence of 1:8000. DM1 is a neglected disease on various levels. No curative treatment exists, and treatment by gene or genetic therapies lies still far in the future. Patients would benefit enormously from treatment of debilitating symptoms like muscle weakness, fatigue and aberrant sleeping behaviour. Metformin and tideglusib are two examples of repurposed drug that are currently in clinical development for DM1, after proven efficacy in preclinical models.
Solution
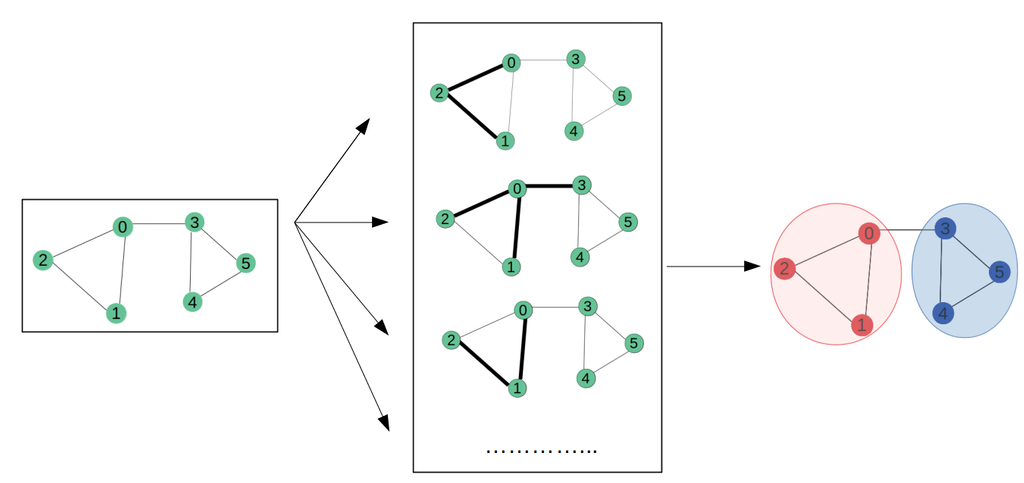
In a previous AI for Health project, we have set up a deep learning framework involving graph neural networks (GNNs) for the prediction of new drug repurposing candidates. A detailed and documented gitlab repository is available. GNNs are particularly suitable for this task because they can exploit a knowledge graph that contains all the known relationships between genes, drugs and diseases. Prediction of new drug repurposing candidates can be defined as a link prediction task, predicting the likelihood of new associations between an existing drug and a given rare disease.
When setting up the GNN framework, we encountered several issues, which we would like to tackle in the current project. (1) the need for a validation strategy. We propose to exploit knowledge graphs that are based on older versions of databases to predict the drugs approved for clinical indications in recent years; (2) the need for providing the underlying evidence for newly predicted associations. We suggest to use explainable AI techniques such as GNNExplainer that present subgraphs of nodes and edges that contribute most to the prediction to the end user; (3) the importance for weighing the different sources of information. Several strategies can be explored here.
Data
You will create different versions of a data repurposing knowledge graph that reflect the discovery of effective drug repurposing strategies over time, by working with historical versions of databases like OpenTargets [INSERT LINK: https://www.opentargets.org] and DisGeNet [INSERT LINK: https://www.disgenet.org]. Lists of successfully repurposed drugs will be extracted from the ReDo database [INSERT LINK: https://www.anticancerfund.org/en/redo-db] and published literature reviews.
Embedding
You will be embedded in the Center for Molecular and Biomolecular Informatics at Radboudumc, and collaborate closely with the Data Science department of the Institute for Computing and Information Systems. We provide access to CPU and GPU clusters, direct supervision by PhD students and regular meetings with the PIs of this project.
Requirements
- Students Artificial Intelligence, Data Science, Computer Science, Bioinformatics in the final stages of their Master education.
- You should be proficient in python programming and have a theoretical understanding of deep learning architectures.
- Basic biological / biomedical knowledge is preferred.
Information
- Project duration: 6 months
- Location: Radboud University Medical Center
- More information can be obtained from Peter-Bram ‘t Hoen (mailto: peter-bram.thoen@radboudumc.nl), Jelle Piepenbrock (mailto: jellepiepenbrock@gmail.com) and Tom Heskes (mailto: tom.heskes@cs.ru.nl)


