This is an AI for Health MSc project. Students are eligible to receive a monthly reimbursement of €500,- for a period of six months. For more information please read the requirements.
Clinical problem
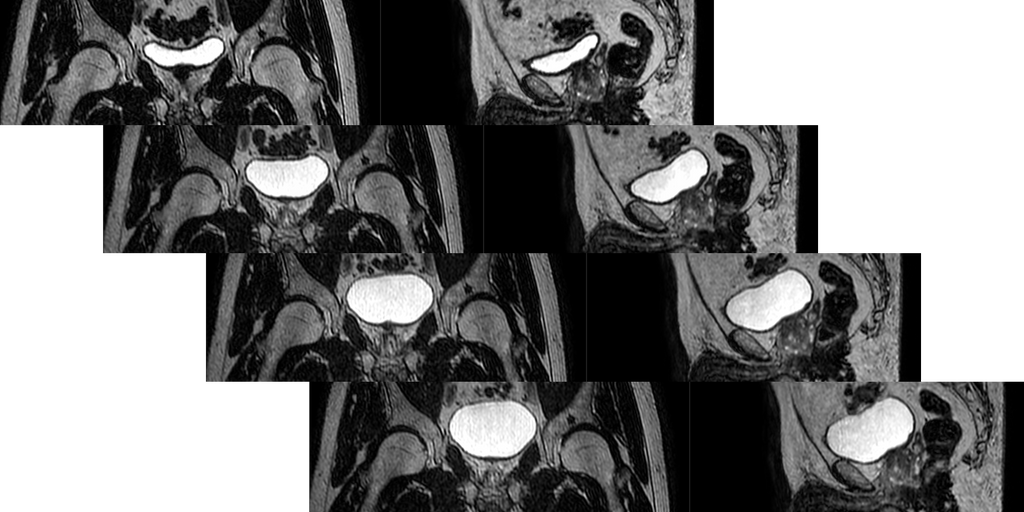
In radiation therapy of cancer, treatment plans are meticulously optimized to maximize the radiation dose in cancerous tissue whilst at the same time minimizing collateral damage to surrounding healthy organs. To this end, prior to treatment, a series of high-quality diagnostic scans are made to precisely delineate target structures and organs-to-be-spared. Subsequently a treatment plan, i.e. the set of instructions passed to the Linac (Linear Accelerator) is created which defines the individualized shapes of the radiation beams. Each individual treatment day (between 3-35 sessions, depending on the tumor type), a localizer scan is made which is used to determine the location of the target on that given day. On conventional linacs this is done with an attached cone-beam CT scanner which offers relatively poor soft tissue contrast, which precludes detailed organ delineation. On MR-Linac systems however (integrated MRI scanner instead of CT), all organs can individually be accounted for very well, hence a better daily treatment plan can be achieved, optimized to the current anatomy. The downside of this procedure is that the time between the localizer scan and the actual start of irradiation is around 15-20 minutes. During this time the bladder naturally continues to fill up which changes the positioning of the nearby structures adversely affecting precision in treatments of cancers near the bladder (pelvic, prostate, etc). And of course, bladder filling continues after the start of irradiation. If deviations from the treatment plan's positioning become too large, treatment has to be stopped prematurely and a new plan has to be calculated which again takes a lot of time.
Solution
Approaches to manipulate bladder filling (e.g. drinking instructions, etc.) have not yielded satisfactory results. As an alternative, being able to predict at what point the change in anatomy will have become too large for effective treatment would be very useful. This could A) indicate how likely it is that intervention will be needed, and B) allow preemptive planning of an alternative treatment on the predicted position such that additional waiting time is minimized. In the current workflow, two MR images are made prior to treatment, one as a localizer at the start which is used for planning, and the second one is made after planning has been completed to enable last-minute rigid-body positional adjustments. As these are approximately 15-20 minutes apart, bladder filling is nicely visualized and these images could potentially be used to extrapolate and predict at what time the anatomy will have changed too much.
Data
At the time of writing, the dataset consists of 79 patients, each with 5 sessions. In each session 4 MR images have been collected (apart from the aforementioned two, also one midway through the treatment and one at the end). These latter two can be used as training targets. Bladder and other organ delineations are available which could be used to evaluate positional prediction performance
Results
The main result will be an AI algorithm that can predict the shape and position of the local anatomy and the expected deterioration of the treatment, which could trigger a decision to adjust the treatment plan midway. Ideally, the predicted anatomy is accurate enough to preemptively calculate a secondary treatment plan in advance to be able to immediately access it when needed.
Requirements
- Students with a major in computer science, biomedical engineering, artificial intelligence, physics, or a related area in the final stage of master level studies are invited to apply
- Experience with programming in Python and its deep learning frameworks
- Theoretical understanding of deep learning architectures
Information
- Project duration: 6 months
- Location: Radboud University Medical Center
- For more information, please contact Peter Koopmans


