Start date: 01-02-2022
End date: 31-07-2022
Clinical problem
Endometrial cancer is one of the more frequent malignancies in women. An increased thickness of the endometrium and abnormal vaginal blood loss are often the first signs of the development of endometrium cancer and early detection and treatment of precancerous tissue (premalignancy) can prevent the ultimate development of cancer, thus preventing the morbidity and mortality. However, only a small percentage of all biopsies (10%) demonstrate endometrial (pre)malignancy and the majority is either benign or is non-informative and has to be sampled again. As the diagnostic evaluation of mostly benign cases results in substantial workload for pathologists, artificial intelligence (AI) assisted pre-selection of suspicious biopsies could optimize workflow. However, the extracted cell tissue is, highly fragmented and pathologically less informative than, for example, a surgical resection, making it a difficult problem for AI assisted diagnosis. Additionally, there has, to date, been no other study on investigating the feasibility of such a system.
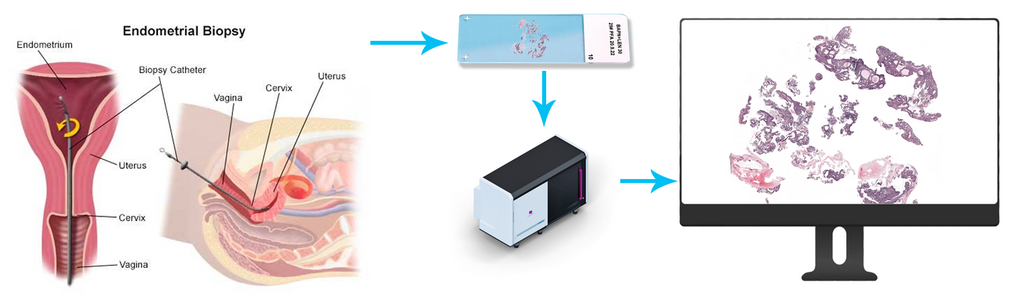
Data
A total of 3002 pipelle biopsies performed between October 2013 and April 2021 were retrieved from the pathology archive of the Radboud UMC. To retain a real-life situation, no preselection of cases was performed. H&E-stained slides were scanned using a 3DHistech P1000 scanner at 0,25 µm/pixel. Images were pseudonymized. A total of 91 whole slide images ( WSI), randomly chosen with a predefined category-weighted key, were immediately set aside to be used in the interobserver variability study and to be used for evaluation of the algorithm. All were given a category by translating the diagnostic report into one of the categories in the table below. 2911 cases were used for training and validating the algorithm.
| Name | Code | Description |
|---|---|---|
| Non-representative | NR | Insufficient tissue for conclusive diagnosis |
| Normal | NL | Cyclical or atrophic endometrium without any signs of pathology or treatment effect |
| Non-neoplastic | NN | Any non-neoplastic change (e.g., treatment effect, endometrial polyp, metaplasia, infection, etc.) which does not belong in any of the other categories |
| Hyperplasia without atypia | H | (possible) Hyperplasia, no mention of atypia |
| Hyperplasia with atypia | AH | (possible) Hyperplasia and (possible) atypia |
| Malignant | M | Any malignancy |
Solution
The initial goal of this study was to create an interpretable algorithm, capable of separating the bulk of non-informative and normal biopsies, from the pre-malignant and malignant ones. This was first extended to a 6-class problem to serve the clinical guidelines, but this did not yield the required performance. The interobserver variability study, performed by AIOS resident pathology Sanne Vermorgen, showed that there was insufficient agreement on certain categories to establish a usable ground truth.
The developed solution is, therefore, an AI-system, based on the multiple instance learning algorithm CLAM[1], that learns to correlate visual patterns on patch-level to binary slide-level diagnosis, based on a single category per WSI.
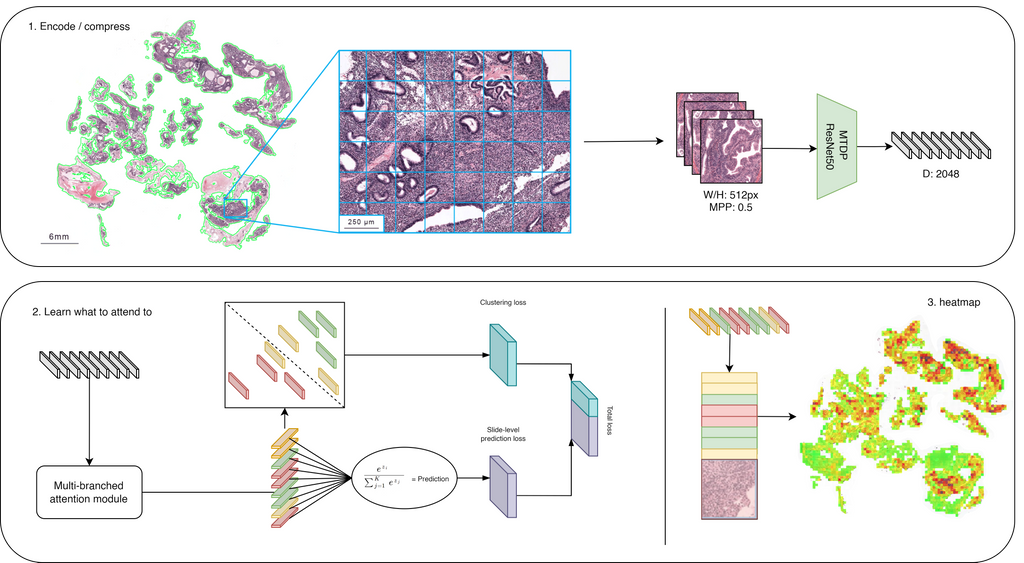
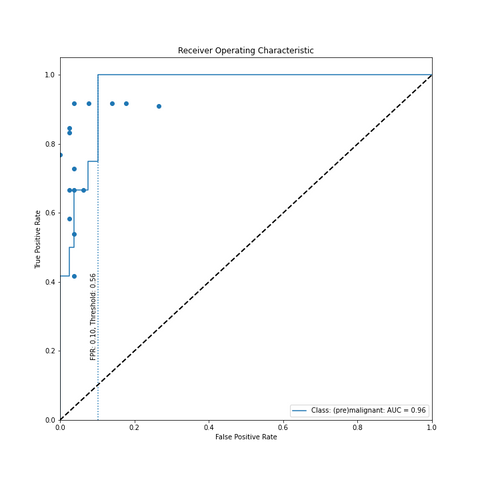
Figure above illustrates the model taking a WSI as input, dividing this into patches (W: 512, H: 512, MMP: 0.5) based on a tissue mask and then encoding each patch into a vector of length 2048. The CNN used for encoding is a ResNet50 architecture pre-trained using 900.000 histopathology images[2]. The vectors are weighted using an attention module and this attention score is used as a pseudo-label. High attention scores should correlate to positive samples and vice versa. These pseudo-labels are used in a clustering module, which could be seen as a form of self-supervised learning and meant to improve the feature representation produced by the attention module. The same attention scores are eventually pooled into a slide-level diagnosis and simultaneously converted into an interpretable heatmap visualisation. The final output of the modul is thus the likelihood that endometrium (pre)malignancy is present and what patches in the image attribute to this. The pathologist can determine whether a WSI is (pre)malignant by determining a threshold themselves. However, a threshold of 0.10 would ensure 100% sensitivity with only a 10% False Positive Rate (FPR). Based on the distribution of the dataset (65% non-(pre)malignant, 35% (pre)malignant) that was used, this means a workload reduction for pathologists of 58.5% (65% - 65% * 0.10).
Conclusion
In this study, we have shown the feasibility of AI-assisted diagnosis for endometrium pipelle biopsies. The fragmented nature of the biopsies, the lack of tissue (and thus pathological information) in most samples and the lack of agreement among pathologists make automation of these screenings a difficult problem. However, when reduced into a binary decision, it shows its value as a tool to potentially be used to pre-screen biopsies and identify all negative cases automatically. The labels were based on the original pathology report, but since the conclusion of that report often consisted of a descriptive diagnosis rather than a definite classification, coding inevitably contained a substantial amount of subjectivity and, therefore, introduced noise. For future studies, the dataset should be curated using the majority vote of multiple coders, which minimizes subjectivity.
Finally, the generalisability of the model should be tested using external cohorts, but the dataset used in this study is the only pipelle sampled endometrium biopsies dataset to date (2022-08-08). Therefore, we encourage medical experts to try the algorithm on grand-challenge by clicking the button below.

Endometrial Carcinoma classification
CLAM model that computes the probability of the WSI being (pre)malignant and also outputs an interpretable heatmap.
Source code can be found on github
Complete thesis on endometrium carcinoma diagnosis using AI techniques is coming soon.
Paper entry with the interobserver variability study for Modern Pathology is coming soon.
References
[1] Lu, M. Y., Williamson, D. F., Chen, T. Y., Chen, R. J., Barbieri, M., & Mahmood, F. (2021). Data-efficient and weakly supervised computational pathology on whole-slide images. Nature biomedical engineering, 5(6), 555-570
[2] Mormont, R., Geurts, P., & Marée, R. (2020). Multi-task pre-training of deep neural networks for digital pathology. IEEE journal of biomedical and health informatics, 25(2), 412-421.
[3] Chen, R. J., Chen, C., Li, Y., Chen, T. Y., Trister, A. D., Krishnan, R. G., & Mahmood, F. (2022). Scaling Vision Transformers to Gigapixel Images via Hierarchical Self-Supervised Learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 16144-16155).


