Clinical Problem
Acute myocardial infarction (MI), also known as a “heart attack”, is a life threatening condition that occurs when blood supply to the heart is decreased or stopped, usually as a result of narrowing / blockage of a coronary artery due to atherosclerotic disease. In patients with MI, coronary angiography (CAG) is usually performed to localize the occluded artery, known as the culprit lesion, and to treat this artery by placing a stent (Dotter procedure). In some cases, more than one lesion is found during CAG. As of yet, it remains largely unknown whether it is beneficial to treat these non-culprit lesions in patients with so-called multi-vessel disease. To date, we cannot accurately predict which atherosclerotic plaques are vulnerable and have a high risk of causing an MI in the future, and which plaques are relatively safe.
Optical coherence tomography (OCT) is a relatively new imaging modality within interventional cardiology and cardiovascular research. OCT uses near-infrared light and is used to provide a high-resolution intravascular image of the coronary artery, allowing for assessment of plaque morphology and characteristics. Previous research has demonstrated its potential on several key aspects in the management of patients with atherosclerotic coronary artery disease, leading to beneficial patient outcome. Hence, OCT is a potential game changes in the field of interventional cardiology, as it may serve as a future tool to distinguish ‘high risk’ from ‘safe’ plaques in patients with acute MI and multi vessel disease.
Currently, however, the use of OCT in daily clinical practice is limited due to a lack of training for assessment and analysis of OCT-acquired pullbacks. In the current project, we aim at easing the use of OCT by means of artificial intelligence (AI), thereby increasing the availability of AI-based OCT algorithms for clinical decision making, and eventually improving patients’ health.
Solution
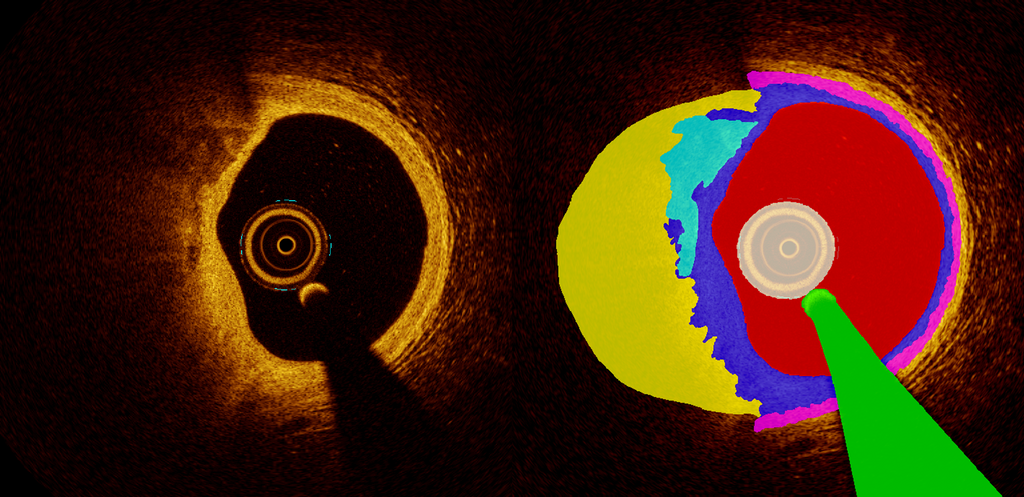
Our group already has developed a prototype algorithm for the segmentation of intracoronary OCT images (Figure 1). Within the current project, we aim to improve this prototype in terms of segmentation of frequently occurring structures (e.g. lumen, vascular layers, lipid, calcium), to improve detection of rarer structures, and to identify markers of plaque vulnerability. Main scientific challenges lie in the development of efficient annotation strategies for individual frames (per-frame analysis) as well as for full pullbacks (multi-frame analysis). Furthermore, AI related challenges include the development of methods to reject unreliable results (outlier classes) and to perform under low-data regime.
To extend current ability of OCT-integrated AI systems to detect luminal border, calcium and the external elastic membrane, we aim to develop a semantic segmentation deep learning algorithm for automated image annotation and target automated segmentation of the lumen, intima, calcium, lipids, media and measures of plaque vulnerability and related complications. The segmentation algorithm will be based on a combination of generative models and semi-supervised learning to address performance in low-data and -annotation regimes. To identify unreliable segmentations, uncertainty-aware predictions will be achieved by model assembling at different granularity levels, and compared to a recently proposed efficient model based on deep deterministic uncertainty.
Data
We will use OCT data from the PECTUS-obs study (https://pubmed.ncbi.nlm.nih.gov/34233996/) to develop and internally validate the algorithm. In total, this database includes 498 intracoronary OCT pullbacks obtained from patients with an acute myocardial infarction and multi-vessel disease. Each pullback consists of 540 frames, adding up to a total of 268,920 individual frames. Currently, the annotation procedure if as follows. All OCT-frames are divided into a training and validation set (ratio 9:1). Manual annotation is performed on every 40th sample of the training set, and supplemented with frames on which “schoolbook examples” of specific structures are visible. Of the training set, 108/498 pullbacks are currently annotated. External validation will be performed on several other prospective databases including high-risk and low-risk populations.
Approach
Students will be supervised by a team with cardiology and deep learning expertise. For model development, we provide access to our deep learning GPU cluster Sol.
Requirements
- Students Artificial Intelligence, Data Science, Computer Science, Bioinformatics, Biomedical Engineering or similar in the final stages of their Master education.
- You should be proficient in Python programming and have a theoretical understanding of deep learning architectures.
- Experience with medical images is beneficial.
Information
- Project duration: 6 months
- Location: Radboud University Medical Center
- For more information, please contact Jos Thannhauser





