Start date: 05-09-2022
End date: 28-02-2023
Clinical problem
Cerebral small vessel disease (CSVD) is a common disorder of the cerebral small vasculature that leads to white matter hyperintensities (WMH), among several other MRI indicators, such as lacunes, enlarged perivascular spaces, cerebral microbleeds, recent small subcortical infarcts, and brain atrophy. CSVD and, therefore, WMH, are commonly seen in elderly subjects and patients with several neurological and vascular disorders. CSVD causes about a fifth of all stroke cases worldwide and is the primary vascular cause of cognitive decline and dementia.
WMH are amongst the foremost recurrent features of CSVD observed on MRI scans. These white matter lesions represent different degrees of axonal loss, demyelination, and gliosis in the brain, which are thought to be caused by a variety of pathological processes, such as inflammation, ischemia, and oxidative stress. WMH have been found to have a strong correlation with common cardiovascular risk factors, specifically hypertension.
WMH appear bright on fluid-attenuated inversion recovery (FLAIR) images and dark on T1-weighted images because these sequences are designed to highlight different tissue characteristics in the brain, specifically the water content of the tissue. While there is evidence linking WMH to gliosis, myelin loss, and axonal loss, the specific mechanisms and pathways involved are still not well understood. It is unclear whether these changes are exclusively related to myelin loss, as WMH can also represent edema. Further research was needed to elucidate the complex and heterogeneous processes underlying WMH development and progression.
In this study, we used the FLAIR and T1 modalities of MRI imaging together with immuno-histochemical (IHC) stainings, specifically Luxol fast blue (LFB), to identify the mechanisms preceding the progression of WMHs in cerebral small vessel disease. IHC is a widely used technique in pathology that allows for the visualization of specific proteins and cellular structures within tissue sections. LFB, in particular, is a commonly used stain that is specific for myelin, allowing for the visualization of myelin loss within tissue sections.
Our study focused on investigating the transition zones from WMH to normal-appearing white matter (NAWM), areas at risk for WMH. By combining MRI imaging with IHC staining, we aimed to elucidate the main contributors to increased WMH burden and its clinical outcome. Specifically, we sought to determine the underlying pathology and mechanisms leading to WMH progression, which is crucial for understanding the pathogenesis of CSVD and developing effective treatments for patients with CSVD.
Overall, our study represents an important step towards understanding the complex and heterogeneous processes underlying WMH development and progression in CSVD. By combining MRI imaging with IHC staining, we were able to gain valuable insights into the pathology and mechanisms leading to WMH progression, which could have significant implications for the development of new treatments for patients with CSVD.
The two main research questions that will be explored in this project are: 1. How can we develop an automatic segmentation pipeline for identifying and segmenting WMH in MRI scans? 2. How can we identify the transition zones between NAWM and WMH?
The goal was to create a segmentation pipeline that can accurately identify and classify WMH and the transition zones between NAWM and WMH, in order to gain a better understanding of the underlying mechanisms behind WMH and their progression. By using high-field MRI and IHC stainings, we can study the relationship between radiological and pathological changes in the brain, and identify potential therapeutic targets for reducing the impact of WMH on brain function.
Data
In vivo MRI
For our study on developing an automatic segmentation pipeline for identifying and segmenting WMH in MRI scans, we used the Radboud University Nijmegen Diffusion Tensor and Magnetic Resonance Imaging Cohort (RUN DMC) data. The data included MRI images acquired at three different time points, all including whole brain scans using 3D T1 magnetization-prepared rapid gradient echo (MPRAGE) imaging and FLAIR pulse sequences. We used semi-supervised labels from a previous study on automated detection of WMH on this dataset, resulting in a dataset of over 1000 cases, from which we used 641 cases after removing unusable cases with bad preprocessing results. The dataset included T1-weighted, FLAIR MRI images, and semi-supervised labels of WMH.
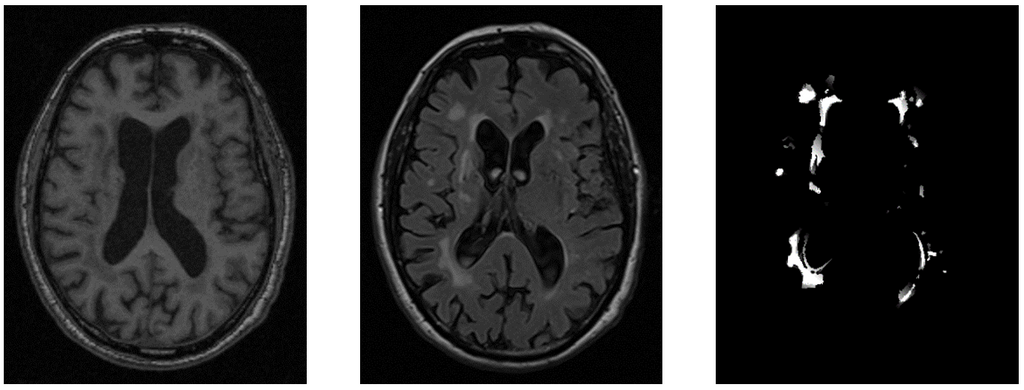
Post-mortem MRI
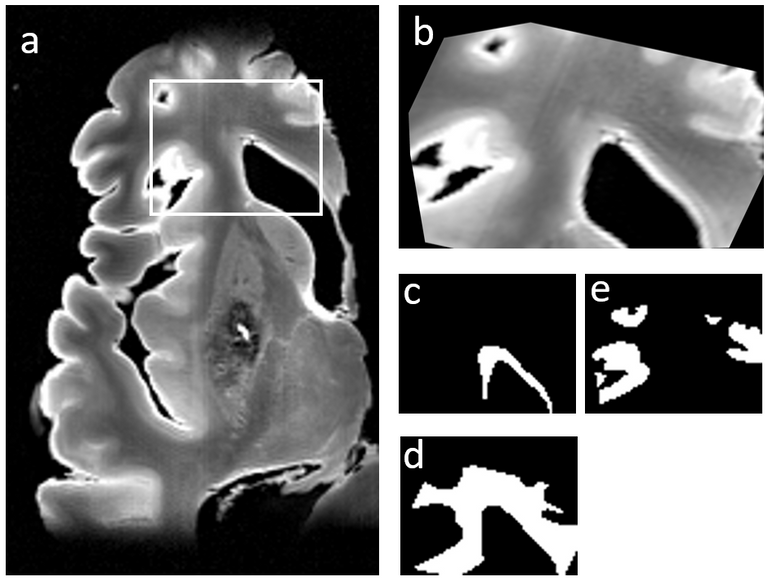
For the second research question, we used a post-mortem dataset of 22 human brains with CSVD radiological hallmarks, including WMH. The brains were horizontally divided into dorsal and ventral parts and scanned using 2D T1-weighted and FLAIR sequences. Semi-automated volumetric segmentation was used to identify WMH, while manual segmentation was used for NAWM and GM. The final labels were based on the agreement of three independent raters.
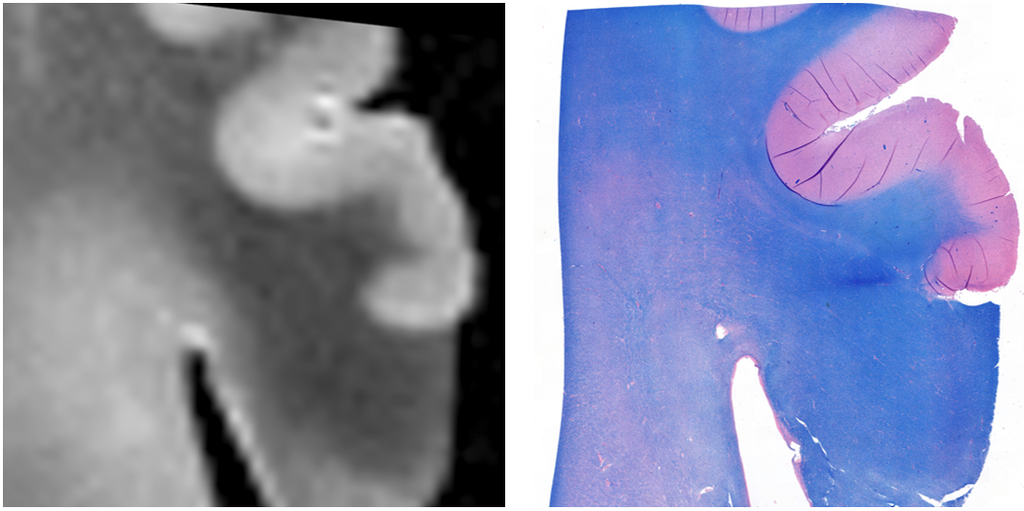
(Immuno-)histochemistry stainings
Tissue blocks from post-mortem brains were sectioned and stained with LFB to detect myelin. Myelin pallor was evaluated as an indirect measure of demyelination. FLAIR MRI data was registered to the LFB reference section using manual landmark selection.
Methods
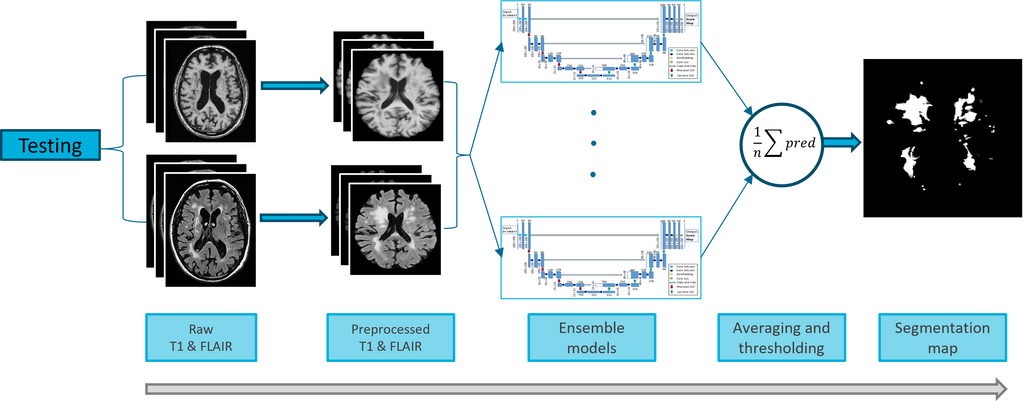
Preprocessing is a crucial step in analyzing medical imaging data. In this study, FSL was used to preprocess MRI FLAIR, T1, and WMH labels. Preprocessing included mapping T1 to FLAIR, removing non-brain tissue, and mapping to MNI-152 standard space. After that, images were normalized, and WMH label values were thresholded to binary. For post-mortem MRI, T1 was mapped to FLAIR to ensure they were in the same coordinate space (using FSL). In total, 541 in vivo MRI cases were used for training, and a specific range of axial slices, between 39 and 149, were selected for processing, resulting in 60,051 images for training. IHC stainings were resized to MRI resolution and background noise was removed.
This study utilized three 2D ensemble U-Nets, a fully convolutional neural network (FCN) architecture, for the segmentation of MRI scans. The U-Net architecture used in this study was based on the winning entry of the WMH Segmentation Challenge, and incorporated techniques and parameters that have been demonstrated to be effective in the winning method of the challenge, with the introduction of dropout and batch normalization layers after each convolutional layer. For in vivo MRI scans, a binary U-Net was used for WMH segmentation, while a multiclass U-Net was employed for WMH, NAWM, and GM segmentation in post-mortem MRI scans. Both models used a batch size of 16, a dropout rate of 0.1, the (weighted categorical) dice loss function, and the Adam optimizer with a learning rate of 2e-3 for optimization. The input shape for the in vivo model was (182, 218, 2), and for the post-mortem model, it was (200, 200, 2).
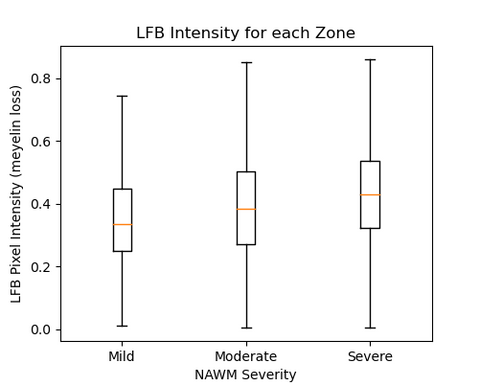
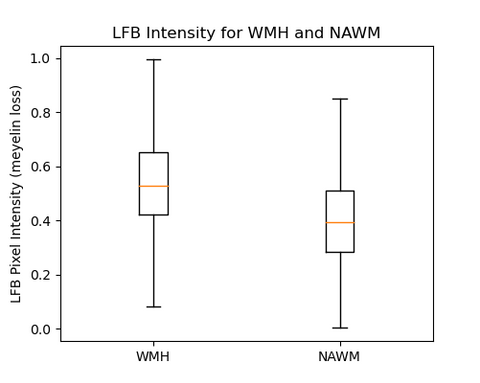
We investigated the progression of WMH and NAWM using FLAIR and T1 weighted images. Prior to creating transition zones, accurate segmentation of WMH, NAWM, and grey matter was necessary. Three transition zones were created within the WMH and NAWM segmentations using KMeans clustering algorithm. A correlation analysis was performed between the MRI data and LFB staining results to study the underlying pathology of WMH. The Spearman rank-order correlation coefficient was used to analyze the correlation between the MRI clusters and LFB staining intensity. LFB intensity values for each cluster were obtained by extracting voxel intensity values and correlating them with the corresponding LFB intensity values.
Results
The model used for in-vivo MRI WMH segmentation has shown good convergence and generalization performance, with a training curve reaching a peak Dice value of 0.76 and a validation curve reaching a peak Dice value of 0.77, compared to 0.805 for human observers. The model can accurately identify WMH and has practical applications. In most cases the model outperforms the labels. This is because the labels are semi-automatic, resulting in not correct labels. Therefore we can not 100% rely on the Dice metric here.
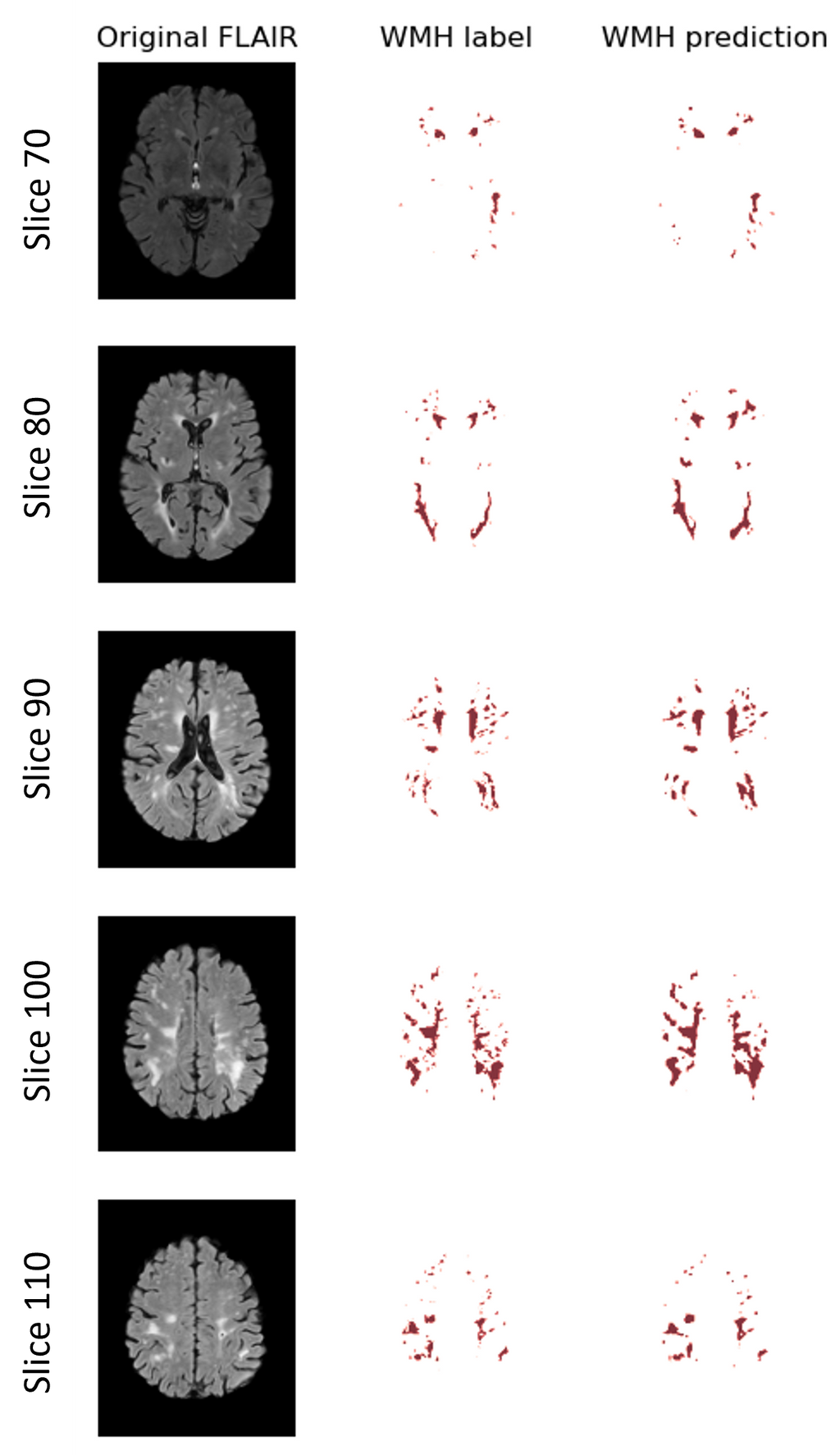
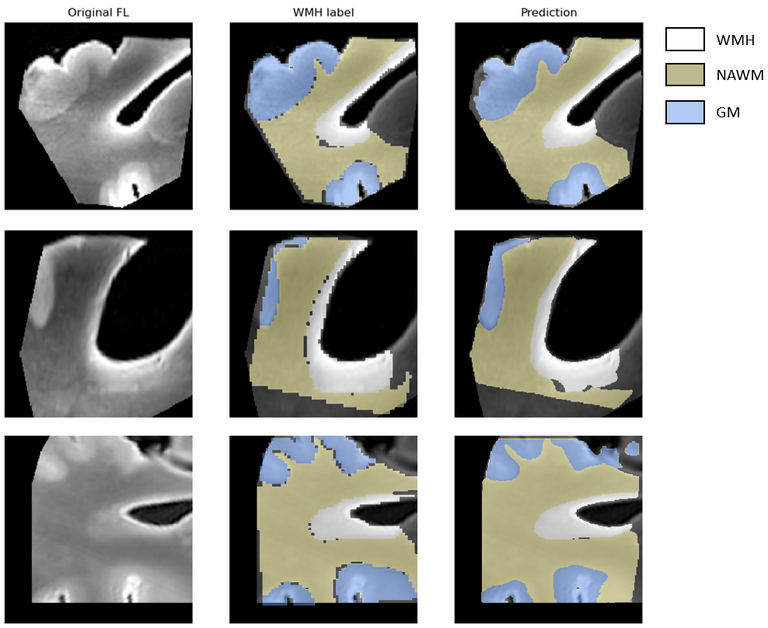
The models used for post-mortem MRI periventricular WMH, NAWM, and GM segmentation show good performance in terms of their ability to correctly segment the target structures in the MRI data, with test Dice coefficients around 0.942. The model can differentiate between the three regions and has practical applications in medical imaging.
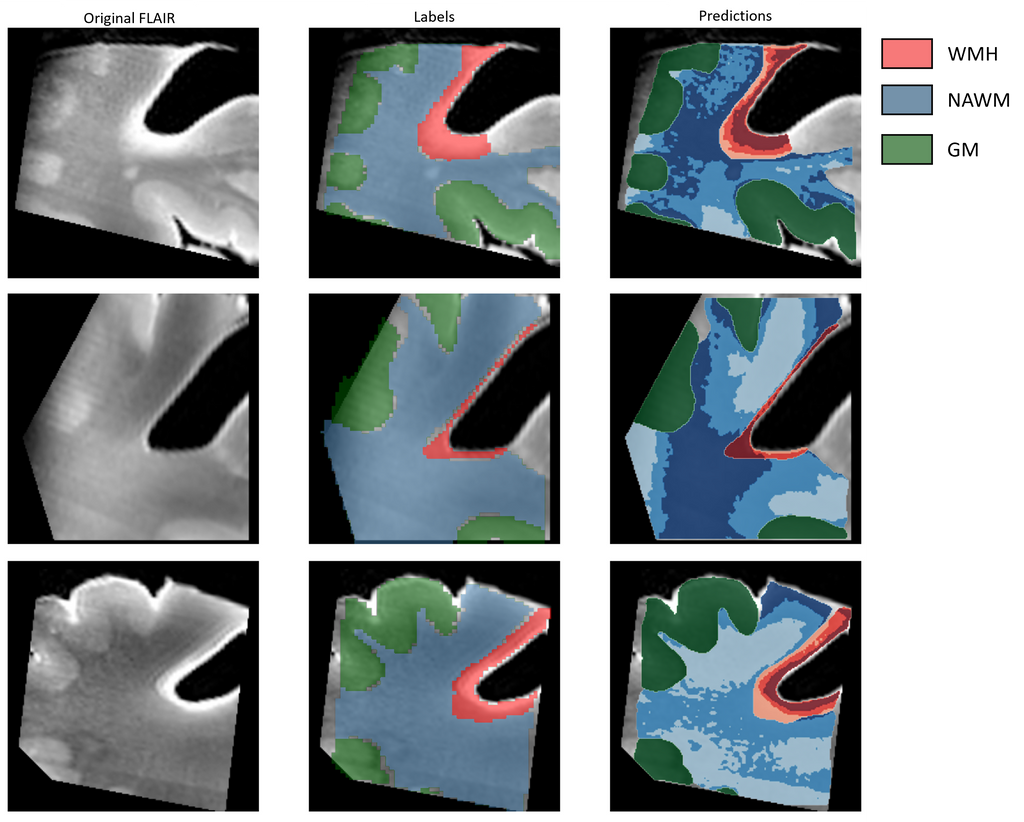
The method used can accurately capture the transition zones between WMH and NAWM, providing a more detailed understanding of the underlying structures in the MRI data. The model can also segment GM in the images but is excluded from the visual representation for transition zones in this analysis.
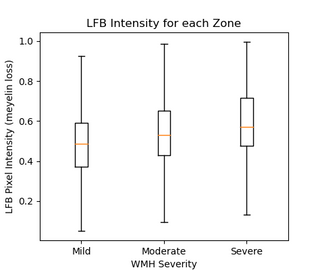
We analyzed the correlation between clustering of WMH and NAWM regions and myelin loss using LFB staining pixel intensity. We found a significant positive correlation, with correlation coefficients of 0.25 and 0.16 for WMH and NAWM, respectively (p-values < 0.0001). We also combined the data for WMH and NAWM, resulting in a correlation coefficient of 0.12 (p-value < 0.0001). These results suggest a positive relationship between the clustering of WMH and NAWM regions, as well as between WMH and NAWM as a whole, and the degree of myelin loss measured by LFB staining.
Conclusion
The study evaluated a proposed method for segmenting WMH in MRI data using Dice coefficient as a performance metric. Results showed high performance compared to human observers. Another experiment segmented three tissue classes using a weighted categorical Dice coefficient, with similar high performance. The study also used k-means clustering to distinguish between different levels of FLAIR intensity in post-mortem MRI data. The study provides new insights into the mechanisms of WMH and their relationship with myelin loss, which has implications for developing therapies to slow or halt progression of related disorders. Future research could explore the relationship between WMH and other markers of white matter damage and perform larger studies on WMH pathogenesis.



