Start date: 01-08-2022
End date: 30-04-2023
Clinical problem
Lung cancer is the leading cause of cancer death among both men and women, accounting for nearly 25% of all cancer deaths. While early stage lung cancer typically shows up as pulmonary nodules on CT examinations, most nodules are benign and do not require further clinical workup. However, radiologist workload is expected to increase soon with the widespread implementation of lung cancer screening programs. Therefore, accurate detection and characterization of pulmonary nodules is crucial to optimize screening.
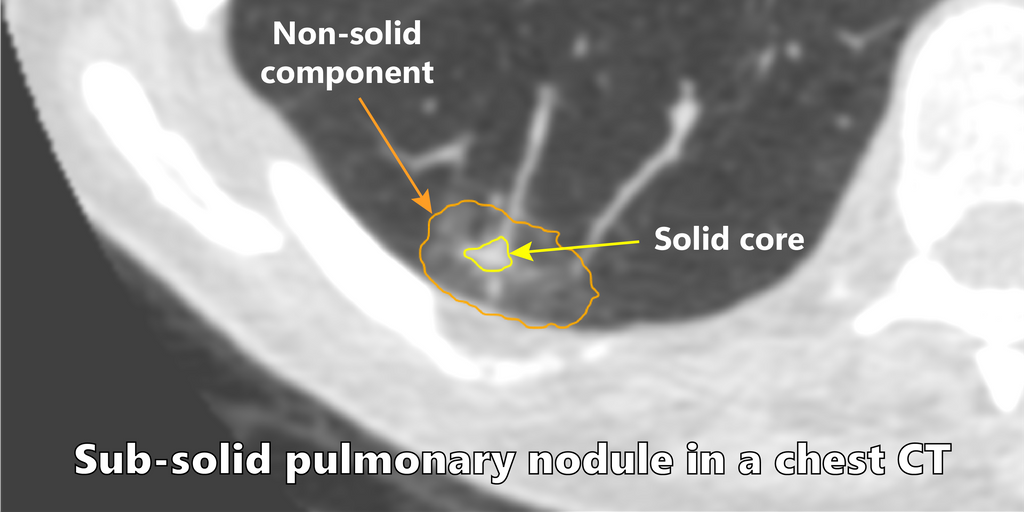
Among the different types of nodules, subsolid pulmonary nodules are encountered in screening and carry a higher malignancy risk. Clinical reporting guidelines recommend different management strategies based on the radiological appearance and biological behaviour of nodules. For subsolid nodules, the management decisions are dependent on accurate volumetric measurements and tracking the evolution of the solid core of these nodules.
Solution
Two nnUNet models were trained to segment the two sub-categories of subsolid nodules, i.e part-solid nodules (PSN) and non-solid nodules (NSN). The algorithm takes a nodule patch as an input and predicts a segmentation mask. An ensemble of 5 nnUNet models is used to generate the segmentation mask.
Approach
To train a network that can predict a segmentation mask, state of the art convolutional neural network approaches were reviewed and adapted to the problem at hand.
Data
A dataset was curated from the MILD cohort. The segmentation mask for the nodules were generated using the CIRRUS lung screening software. An expert radiologist finetuned the segmentation mask obtained from the CIRRUS software. A visual check was performed on the nodules to exclude nodules which contained bad segmentations (e.g. scarring was segmented, vessels were segmented as solid core).
For the PSN network 132 nodules and for NSN 613 nodules were used for training the network.
For part-solid nodules the segmentation mask contained three classes namely background, non-solid and solid core. For NSN, only two classes were present: background and non-solid region.
The training data consist of patches of size 64x32x32 (z, x, y) with the nodule located at the center of the patch. The patches were clipped using the intensity values of 0.5 and 99.5 percentile of the foreground voxels. Each patch was then normalized over the mean and s.d. of intensity values of entire dataset. On the fly augmentations were performed to enlarge the dataset for training.
The DLCST cohort was used as an external dataset to validate the network accuracy on unseen data. This contained 119 PSN and 105 NSN nodules. The segmentation reference masks for the nodules were again generated using the CIRRUS software and finetuned by the radiologist.
Results
The segmentation network for part-solid network achieved a dice score of 77.6% for non-solid and 76% solid core on the external dataset. For the NSN task the network achieved a dice score of 83.3%. The table below shows the distribution of dice score across different categories defined using the nodule size measurement stated in Lung-RADS guidelines.
Dice scores for part solid nodules
| Lung RADS Category | Count | Non-Solid | Solid Core |
|---|---|---|---|
| Probably Benign | 69 | 79.2% | 76.3% |
| Suspicious | 21 | 80.7% | 79% |
| Very Suspicious | 29 | 71.9% | 73.5% |
| Mean | 119 | 77.6% | 76% |
Dice scores for ground glass opacities
| Lung RADS Category | Count | GGO |
|---|---|---|
| Mean (“Benign”) | 105 | 83.3% |
Conclusion
The current research achieved good results for subsolid nodules. The network is capable to produce robust segmentations and avoid segmentation of vessels and chest-wall. Future research should focus on building a unified network capable of classifying and segmenting all different nodule type in a single shot. A self-supervised approach can be utilised to generate pseudo labels and reannotate the nodules which contained inaccurate segmentation. The class imbalance issue needs to be addressed since solid nodules are more frequent when dealing with all nodule types. The nnUNet framework is suboptimal when encountering extreme class imbalance and did not produce good results when the dataset for all nodule types are used.
The final thesis report has more details about the project and experiments performed.
An interactive demo is accessible via Grand-Challenge: Try out the algorithm
The code for the project can be found in the GitHub Repository


