Start date: 30-09-2020
End date: 01-04-2021
Clinical problem
In a supervised learning setting, we need labeled data in order to train a machine learning model. The acquisition of labeled training data is a time consuming and costly task - if done manually. An alternative to hand-labeling training data is to create training data programmatically. Weak supervision is an approach to label training data programmatically, where the user defines multiple labeling functions (LFs). The LFs can differ in type, for example, regular expressions to capture a certain re-occurring structured private information, or use an external knowledge base to find known entities. The user defined LFs are then aggregated to output a probabilistic label to the training data.
In our project, we have clinical records that may contain protected health information (PHI). It is important to annotate and replace PHI in unstructured medical records, before being able to share the data for other research purposes due to privacy legislation. The process of replacing protected health information is also known as de-identification.
There are different approaches to deidentify clinical data: rule-based, feature based and deep neural methods. Rule-based models implement multiple rules to find private information in clinical reports, whereas the machine learning models require training data to train and validate the performance.
Solution
The RUMC Radiology department already employs a rule-based approach, which comprises a complex set of rules to do the de-identification on the radiology reports. Our project contributes to the Radboud Routine Care in two ways:
- First, as there are already hand-labeled training data, we want to experiment with a different approach other than the implemented rule-based method. We want to investigate whether there is any improvement in predictive performance.
- Second, a significant amount of the radiology report still needs to be annotated. More data often leads to better predictive performance, especially in deep neural methods. Therefore, we also want to apply weak supervision to programmatically label the radiology reports to investigate the effect of programmatically generated labels on de-identification of radiology reports.
Data
In this project, we will use the clinical records from the RUMC Radiology department, which are in Dutch.
This dataset comprises over 1600 hand annotated documents.
Eight different PHI tags are defined, but the distribution of the tag count differs.
The distribution of the tag counts are as follow: (Persoon, 129), (Datum, 1833), (Tijd, 82), (Telefoonnumer, 18), (Patientnummer, 0), (Znummer, 0), (Leeftijd, 2) and (Plaats, 5) which puts the total on 2069 tags.
The majority of the tags are datum instances
Approach and Results
Our approach for this project will be as follow:
- As for the first solution, we take the rule-based implementation as baseline and want to experiment with a feature-based method and a deep neural method.
- As for the second solution, we want to experiment with weak supervision for de-identification where we collect training data using labeling functions.
De-identification using human annotated data
The first solution is to implement machine learning models to compare against the current report anonymizer (RRA). We have implemented two different machine learning models. Our first model is a conditional random field (CRF) where we have extracted multiple informative features from the text. Our second model is a bidirectional-LSTM (BI-LSTM) which does not require feature extraction, but rather it learns the internal representations automatically. To evaluate the performance, we report precision, recall and F1 scores. The results for our machine learning models using human annotated data on the RUMC dataset can be found in the table below. Our implementation of de-identification models approaches the performance of the RRA (F1 score of 1.00), but for tags that occur less frequently in the training data, the performance degrades. One solution is to collect more training data.
| Model | Precision | Recall | F1 |
|---|---|---|---|
| RRA | 1.00 | 1.00 | 1.00 |
| CRF | 0.94 | 0.82 | 0.88 |
| BI-LSTM | 0.80 | 0.75 | 0.77 |
De-identification using weakly tagged data
We can collect training data by doing it manually, but this is a repetitive and time-consuming task. Our second solution is to apply weak supervision to collect and annotate training data. The idea of weak supervision is to write multiple labeling functions which are simple programs that output a label. Subsequently, the labels from the different labeling functions are aggregated (we used the Snorkel framework to apply weak supervision). The results for our machine learning models using weakly annotated data on the RUMC dataset can be found in the table below. We see that our CRF implementation with weakly tagged training data approaches the performance of human annotated dataset (F1 score of 0.76 and 0.88, respectively).
| Model | Precision | Recall | F1 |
|---|---|---|---|
| RRA | - | - | - |
| CRF | 0.75 | 0.76 | 0.76 |
| BI-LSTM | 0.61 | 0.72 | 0.66 |
Through manual inspection of the results, we note that for structured entities, such as datum and tijd, we obtain near similar F1 scores, but entities that have numerous ways (e.g. persoon) to express need additional attention.
| Entity | Gold F1 | Weak F1 |
|---|---|---|
| Datum | 0.92 | 0.80 |
| Tijd | 0.67 | 0.80 |
| Persoon | 0.48 | 0.18 |
Additionally, we experimented to see the effect of increasing the training data size on the predictive performance, because entities that occur less frequently had a lower performance. In the figure below, we increased the training data using different percentages of the weakly tagged training data and we see that after 40 percent of the data, the curve flattens for our implementation of CRF using weakly tagged training data.
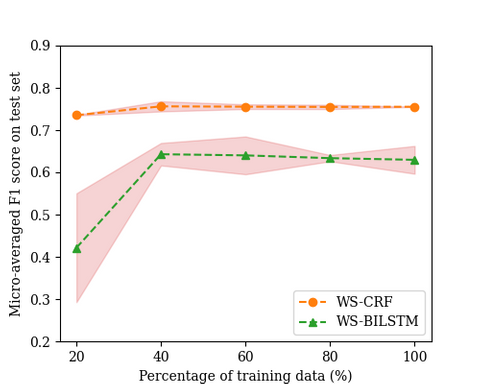
However, if we look at each entity individually, we see that adding more instances of datum has minimal impact, whereas the persoon instance has a gradual increase but the performance lacks.
In the table below, we display the effect of increasing the training data size per entity on the performance using the F1 score.
| Entity | 20% | 40% | 60% | 80% | 100% |
|---|---|---|---|---|---|
| Datum | 0.79 | 0.81 | 0.80 | 0.80 | 0.80 |
| Tijd | 0.24 | 0.62 | 0.74 | 0.80 | 0.80 |
| Persoon | 0.06 | 0.07 | 0.10 | 0.14 | 0.18 |
Conclusion
In conclusion, we have implemented de-identification models using machine learning models which are quick to implement, but require training data.
Collecting and annotating training data is a time-consuming task, thus it is important to study how we can automate this process.
In this project, we applied weak supervision to acquire training data, where we see that for structured entities in the problem of de-identification, we can achieve competitive results.
However, more complex entities require more attention as capturing all the varities, for instance for persoon instance, is a challenging task.
The code and report for this project can found in this Github repository.


