Start date: 01-02-2022
End date: 31-07-2022
Clinical Problem
The development of novel drugs is an expensive, time-consuming and risky process. Drug repurposing, where already tested and approved drugs are used for other indications than they were originally developed for, can be a possible solution to this problem. Drug repurposing can significantly shorten the development and testing process, decreasing developmental costs, duration and the risk of testing involved. This especially can be of great value for novel and rare diseases. This project presents a method that uses graph neural networks for link prediction on biomedical knowledge graphs in order to predict potential drug repurposing candidates. We focussed on the prediction of drug repurposing candidates for the novel COVID-19 disease and the rare disease Myotonic Dystrophy type 1 (DM1). The outbreak of COVID-19 led to a global pandemic in 2019. The contagious disease is caused by the SARS-CoV-2 virus. Its common symptoms are e.g. fever, cough and fatigue. More severe symptoms such as breathing difficulties can lead to hospitalization. DM1 is a rare, genetic, muscular disease that affects at least 1 in 8,000 people. DM1 is highly variable in severity, type of symptoms and the age of onset. Its common symptoms are muscle weakness, muscle stiffness, daytime sleepiness and disturbance of the heart rhythm. For both diseases, there is no effective treatment yet. Therefore, drug repurposing using graph neural networks can be of great value for these diseases.
Methods
In this study, the drug repurposing knowledge graph (DRKG) is used as the biomedical knowledge graph. This knowledge graph incorporates knowledge on many different diseases, including COVID-19 and DM1, represented in a triple format. For example, [drug, treats, disease].
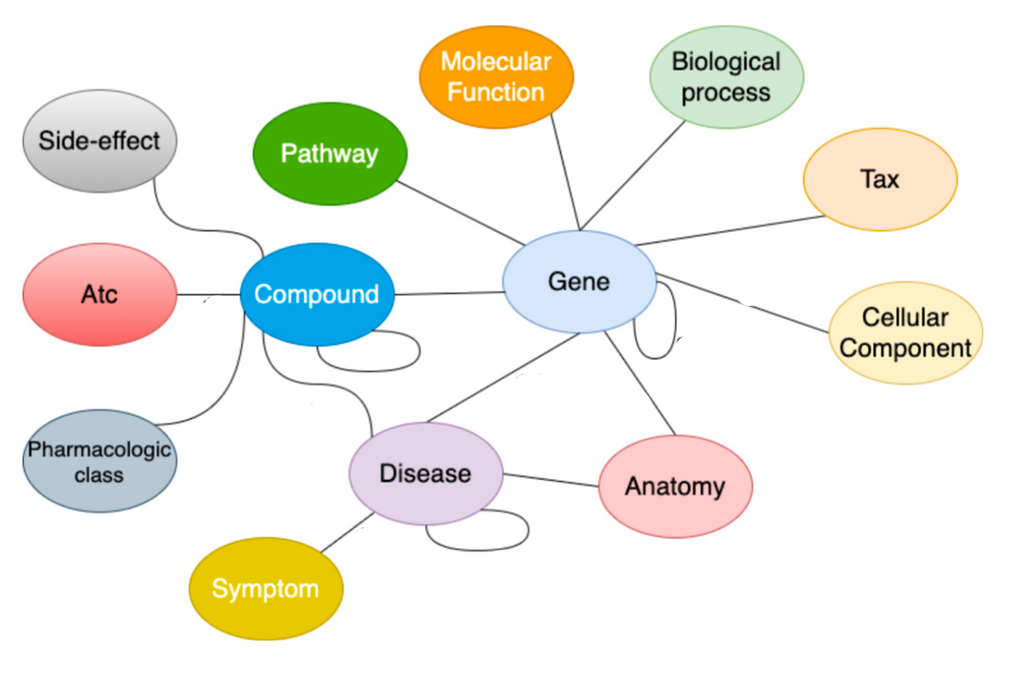
To find drug repurposing candidates, a relational graph autoencoder (GAE) is implemented to learn a low-dimensional representation (embedding) of the knowledge graph. The model is optimized using balanced batches (i.e., minority classes are oversampled to account for edge-type imbalance) with a binary cross-entropy loss. Each node embedding is learnt using the local structure of the neighbourhood of a node. Potential drug repurposing candidates are computed by a link prediction task, where the similarity between two node embeddings of interest is computed. Nodes are likely to be connected when the node embeddings show high similarity. We are therefore looking for drug and disease nodes with similar embeddings that are not yet connected in the original knowledge graph. The embeddings are evaluated using AUROC and AUPRC.
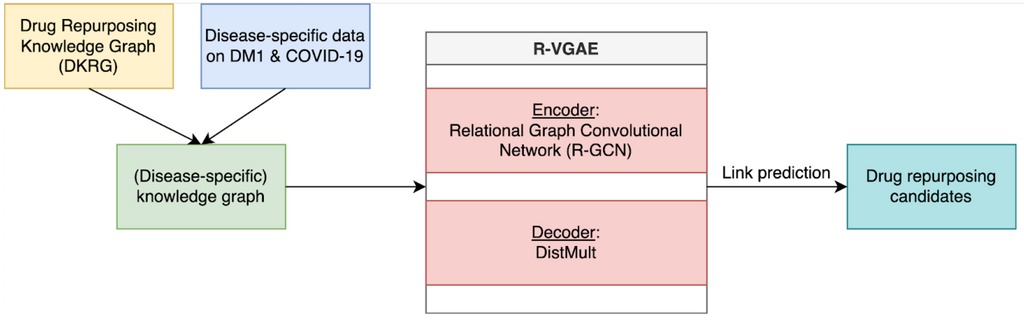
We enriched the generic DRKG using experimental disease-specific information. For COVID-19, an analysis on transcriptomic data of hospitalised patients was conducted to identify specific gene-gene relations that were added to the network. For DM1, an analysis on transcriptomic data measured in the brain, blood and muscles was used to identify relevant genes and gene-gene relations. Additionally, we experimented with knowledge directly linked to the disease of interest. For COVID-19, we used novel knowledge obtained from the fast-growing amount of research. For DM1, we used a dataset on symptoms and associated genes. We investigated how the predictions of these enriched knowledge graphs differed from the predictions made using the generic knowledge graph and whether this leads to more relevant results.
Results
As we investigated 2 diseases, the results will be described for both diseases below. A more in-depth analysis can be found in the theses.
COVID-19
Generic knowledge graph
The table below shows the top-10 ranked drug repurposing candidates for COVID-19 based on the generic knowledge graph. As you can see, there are mostly generic and irrelevant drugs in this top-10.
| Drugname | Probability | Out-degree |
|---|---|---|
| Cyclosporine | 0.989 | 2924 |
| Colchicine | 0.987 | 964 |
| Methotrexate | 0.982 | 2030 |
| Cyclophosphamide | 0.979 | 1435 |
| Cisplatin | 0.978 | 1868 |
| Simvastatin | 0.976 | 1569 |
| Paclitaxel | 0.975 | 1505 |
| Curcumin | 0.974 | 544 |
| Doxorubicin | 0.974 | 2011 |
| Cholesterol | 0.974 | 854 |
Disease-specific gene-expression knowledge graph
After adding the gene-expression data, the following top-10 is obtained. Again, we see mostly generic and irrelevant drugs. One interesting drug in this top-10 is dexamethasone. This is an anti-inflammatory that is FDA approved as a COVID-19 treatment. It is found that this treatment reduces ventilation and oxygen need. However, there is some overlap with the top-10 obtained from the generic knowledge graph. Therefore, we looked at the differences between predicted probabilities of all drugs for both knowledge graphs, shown in the figure below.
| Drugname | Probability | Degree |
|---|---|---|
| Cyclosporine | 0.991 | 2924 |
| Clarithromycin | 0.987 | 783 |
| Methotrexate | 0.980 | 2030 |
| Ketoconazole | 0.978 | 874 |
| Fluconazole | 0.977 | 1503 |
| Dexamethasone | 0.977 | 2158 |
| Azithromycin | 0.974 | 1187 |
| Itraconazole | 0.974 | 806 |
| Prednisolone | 0.974 | 1135 |
| Dextrose, unspecified form | 0.973 | 1691 |
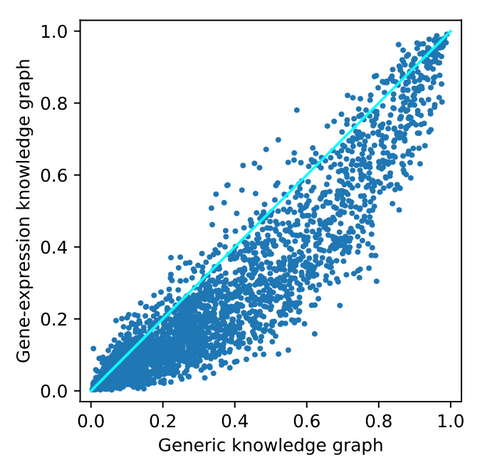
From this image, it can be observed that there is a small effect on some of the drugs. Most drugs are pushed to the bottom of the ranking. However, there are also some drugs pushed up the ranking. These are potentially interesting as they are positively affected by adding COVID-19 related data. For a closer look into these drugs, we refer to the thesis.
New direct evidence knowledge graph
From adding new direct evidence, the following top-10 drug repurposing candidates are obtained. As seen before, there are mostly generic and irrelevant drugs observed here. Again, cyclosporine is ranked as the best drug repurposing candidate. However, from the figure below, it can be observed that the drugs are substantially more affected by adding the new direct evidence. This implies that the effect of adding new evidence that is directly linked to the disease node has a bigger effect on the ranking. For a closer look into these affected drugs, we refer to the thesis.
| Drugname | Probability | Degree |
|---|---|---|
| Cyclosporine | 0.985 | 2924 |
| Clarithromycin | 0.975 | 783 |
| Itraconazole | 0.973 | 806 |
| Fluconazole | 0.970 | 1503 |
| Zidovudine | 0.966 | 1127 |
| Colchicine | 0.965 | 964 |
| Ketoconazole | 0.960 | 874 |
| Azithromycin | 0.956 | 1187 |
| Methotrexate | 0.955 | 2030 |
| Omeprazole | 0.954 | 1215 |
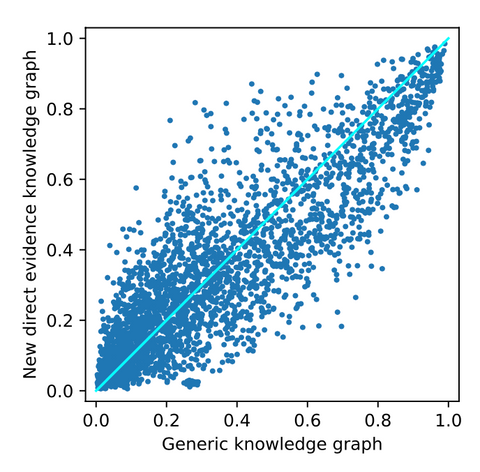
The results above show that the biggest effect is observed when the generic knowledge graph is enhanced with novel knowledge that is directly linked to the disease node of interest. Although a bigger difference does not imply better results, there is at least more effect from adding this data.
DM1
In the table below, the top-10 predicted drugs using the generic DRKG can be found. Norepinephrine, Epinephrine & Dopamine at first seemed interested as they are linked to the treatment of symptoms related to DM1. However, in practice, they are only used in severe cases which makes them unlikely drug repurposing candidates. All other drugs are likely to be irrelevant.
| Drug name | Probability | Out-degree |
|---|---|---|
| Ethanol | 0.992 | 1637 |
| Dextrose | 0.990 | 1691 |
| Oxygen | 0.983 | 967 |
| Cholesterol | 0.982 | 854 |
| Norepinephrine | 0.978 | 876 |
| Epinephrine | 0.977 | 992 |
| Melatonin | 0.977 | 1038 |
| Dopamine | 0.977 | 1047 |
| Ascorbic acid | 0.976 | 707 |
| Curcumin | 0.976 | 544 |
Extending DRKG with disease-specific gene-gene edges
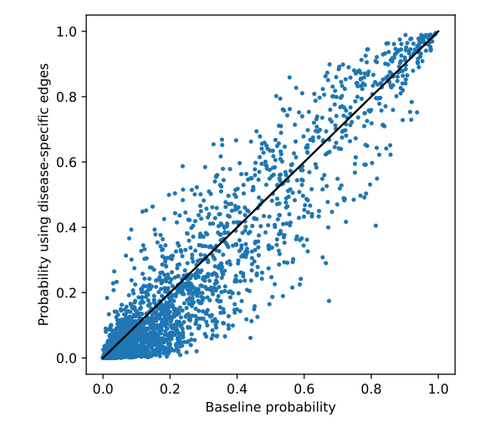
In the first series of experiments, DRKG was enriched with disease-specific gene-gene edges derived from the aforementioned gene-expression datasets. It was observed that adding these edges only had a limited impact on the ranking (i.e., the probabilities and ranking positions of the drugs did not change a lot). Below, the top-10 predicted drugs using the DRKG extended with DM1-related gene-gene edges derived from blood gene-expression data can be seen. This ranking differed most from the ranking predicted using the generic knowledge graph. 4 drugs are overlapping with the generic ranking. Metformin is an interesting prediction as it showed efficacy for DM1 in preclinical research. However, it is connected via a direct edge in DRKG. All other drugs are either irrelevant or do not show a clear connection to DM1.
| Drug name | Probability | Out-degree |
|---|---|---|
| Ethanol | 0.995 | 1637 |
| Dextrose | 0.990 | 1691 |
| Melatonin | 0.990 | 1038 |
| Nitric Oxide | 0.989 | 1590 |
| Dexamethasone | 0.989 | 2158 |
| Acetylsalicylic acid | 0.989 | 1492 |
| Metformin | 0.989 | 1436 |
| Progesterone | 0.985 | 1768 |
| Nicotine | 0.985 | 1444 |
| Oxygen | 0.984 | 967 |
Below, the predicted probability for each drug in the generic ranking is plotted against the probability predicted in this experiment. As can be seen, probabilities are more similar at the top and bottom than they are in between. These drugs are analysed in detail in the thesis.
Extending DRKG with DM1 related symptoms and associated genes
In the second experiment, DRKG was extended with knowledge on DM1-related symptoms and associated genes. The ranking and probabilities per drug showed substantially more different from the generic ranking compared to the gene-expression experiments as also shown in the scatter plot below. 2 drugs are overlapping with the generic ranking. Phenytoin and Colchicine are interesting predictions. Phenytoin is used for the treatment of Myotonia, which is a symptom of DM1. Colchicine showed efficacy in previous research. Again, for all other drugs, no direct link to DM1 or one of its symptoms was found.
| Drug name | Probability | Out-degree |
|---|---|---|
| Phenytoin | 0.982 | 1642 |
| Colchicine | 0.979 | 964 |
| Dinoprostone | 0.979 | 693 |
| Vitamin E | 0.978 | 480 |
| Dexamethasone | 0.977 | 2158 |
| Acetylsalicylic acid | 0.976 | 1492 |
| Hydrocortisone | 0.976 | 1550 |
| Ethanol | 0.976 | 1637 |
| Epinephrine | 0.975 | 992 |
| Morphine | 0.975 | 1678 |
The scatter plot below shows that the probabilities predicted for each drug differed more from the generic ranking compared to the gene-expression experiments. These drugs are also analysed in more detail in the thesis.
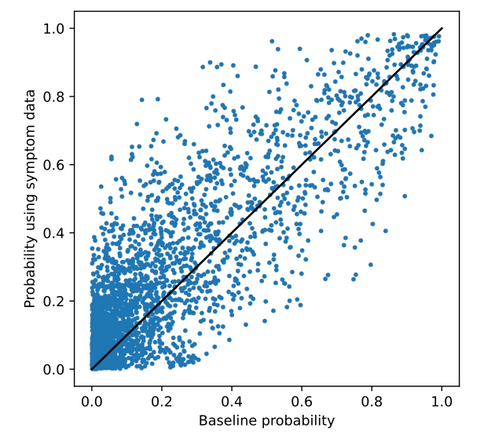
Similar to the results on COVID-19, it can be seen that adding edges directly to the disease in the knowledge graph (knowledge on disease symptoms) results in a more different ranking. However, it did not lead to the prediction of drugs for which it is evident that they are useful for the treatment of DM1. Therefore, a more different ranking again does not necessarily mean a better ranking in terms of potential efficacy.
Conclusion
During this project, we developed a pipeline for the prediction of drug repurposing candidates using graph neural networks. In comparison to published work, we tried to make a more disease-specific knowledge graph. Potentially resulting in more relevant drug repurposing candidates. The results show that there are differences between the predicted rankings for both the diseases that were investigated. However, at a first glance, the top-ranked drug repurposing candidates are mostly generic and irrelevant. Furthermore, the top of the ranking is influenced by highly connected nodes, which introduces a bias in our results towards these highly connected nodes, making the results less reliable. Future work should be aimed at mitigating this bias. The results show that there is a clearer difference in predicted probabilities when enhancing the generic knowledge graph with direct edges compared to indirect edges (i.e. the gene-gene edges). However, it is hard to validate the results as there is no ground truth available as there is no cure for the diseases we investigated.



