Start date: 30-01-2023
End date: 30-07-2023
Clinical problem
Every day, hundreds of histopathology slides are made in our medical center. Most of them are stained with hematoxylin and eosin (H&E) to visualize tissue morphology, producing the typical 'pink-purple-blue pathology slides'. In some cases, specific subtypes of cells are targeted with immunohistochemistry (IHC), where specific proteins can be stained using the specificity of antibodies. In this case, cell nuclei are usually stained in blue, and targeted cells are stained using a different chromogen (e.g., DAB in brown), which can vary between laboratories.
Both H&E- and IHC-stained slides are standardly produced at large scale in pathology laboratories all around the world, usually scanned via bright-field microscopy. In recent years, a large corpus of deep learning algorithms has been proposed to automate quantification on H&E- and IHC-stained images, usually trained based on large-scale datasets with manual annotations created by several research groups, including ours. However, the use of different IHC chromogens across laboratories hinders the applicability of one algorithm trained on a particular color to a laboratory using a different color.
At the same time, it is possible to target multiple types of cells on the same tissue sample using more advanced techniques such as multiplex immunofluorescence (mIF), a more expensive, time-consuming, and not largely available, yet powerful research technique that is increasingly used for biomarker analysis and discovery. Despite some recent developments in the field, to date, the analysis of mIF data is mostly based on traditional image analysis models and still does not benefit from the availability of large-scale curated datasets produced for AI development in bright-field microscopy.
Solution
In this project, we developed an approach for making AI models for automated quantification in digital pathology applicable across two domains: standard immunohistochemistry in bright-field microscopy using different chromogens, and multiplex immunofluorescence.
Approach
The core principle of our approach is to replace the RGB input typically used to train deep learning algorithms with a combination of relevant density channels extracted from the image itself, such as the intensity of nuclei and the intensity of the chromogen attached to the targeted protein in IHC. For 'unmixing' RGB images into stain density channels, our approach leverages color deconvolution based on initial work developed in our group (Geijs et al. 2018, 10.1117/12.2293734).
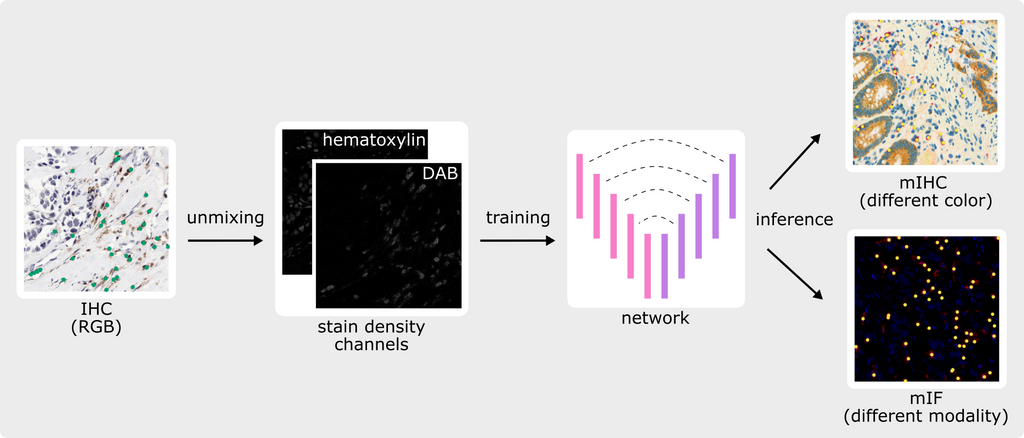
Since the input of the deep learning model is no longer an RGB image but instead pure density channels, the algorithm can be applied to IHC data stained with a different chromogen without the need to train on all possible chromogen variations. Moreover, the same algorithm can be de facto applied to other imaging modalities such as mIF by taking combinations of intensity channels like DAPI for nuclei and specific antibody channels for targeted cells. Without the need for collecting, curating, and annotating new training data for deep learning in mIF, our approach allows the reusing of large-scale digitalized archival material and corresponding annotations to bring deep learning-based quantification to the field of spatial biology and biomarker discovery in mIF.
Data
We used lymphocyte detection in IHC as a use case and reused existing IHC data generated via bright-field microscopy and annotated in past projects. In particular, we used the dataset reported in (Swiderska-Chadaj et al. 2019, 10.1016/j.media.2019.101547), consisting of n=83 DAB-stained IHC images from three different organs (breast, colon, and prostate) and >170,000 manually annotated CD8+ or CD3+ lymphocytes. This dataset includes training and validation sets as well as the public LYON19 test set. For application to differently-stained IHC data, we used a colorectal cancer (CRC) dataset partially described in (Studer et al. 2020), which consists of n=76 multiplex IHC (mIHC)-stained images with >20,000 manual annotations of red-stained CD8+ lymphocytes.
For application to mIF data, we used two datasets with markers for both nuclei and CD8: (1) the PINNACLE (PIN) dataset, consisting of n=18 lung cancer images and >4,900 new manual annotations, and (2) the MSKCC dataset, a selection of 29 patches of various cancers from (Aleynick et al. 2023, 10.1038/s41597-023-02108-z), for which we automatically generated >1,200 annotations based on the CD8 channel and provided whole-cell and nuclei masks.
Results
We applied our approach to a modified version of a published U-Net (Swiderska-Chadaj et al. 2019, 10.1016/j.media.2019.101547) and compared it with an RGB-trained baseline on all four datasets. For a fair comparison, we applied the RGB-trained network to artificially generated bright-field simplex IHC versions of the mIHC and mIF datasets. The table below presents the F1-score, precision, and recall (average ± standard deviation over five experiments) of the RGB- and unmixed-trained networks, and the figure shows the network predictions together with the annotations for one patch of each test set.
| Dataset | Type | Method | F1-score | Precision | Recall |
|---|---|---|---|---|---|
| LYON19 | IHC | RGB | 0.79 ± 0.01 | 0.74 ± 0.02 | 0.85 ± 0.01 |
| Unmixed | 0.78 ± 0.01 | 0.73 ± 0.02 | 0.83 ± 0.02 | ||
| CRC | mIHC | RGBartificial-IHC | 0.82 ± 0.02 | 0.81 ± 0.02 | 0.83 ± 0.04 |
| Unmixed | 0.74 ± 0.01 | 0.63 ± 0.02 | 0.90 ± 0.01 | ||
| PIN | mIF | RGBartificial-IHC | 0.50 ± 0.15 | 0.67 ± 0.06 | 0.45 ± 0.18 |
| Unmixed | 0.62 ± 0.02 | 0.46 ± 0.02 | 0.96 ± 0.02 | ||
| MSKCC | mIF | RGBartificial-IHC | 0.44 ± 0.17 | 0.74 ± 0.31 | 0.32 ± 0.11 |
| Unmixed | 0.72 ± 0.02 | 0.73 ± 0.04 | 0.72 ± 0.05 |
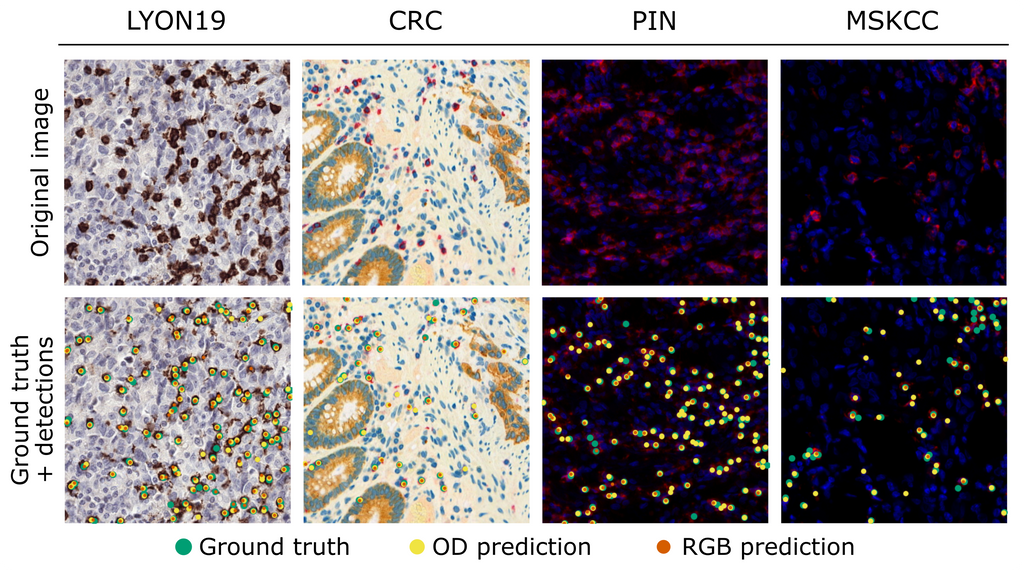
Both networks achieved a similar lymphocyte detection performance on the LYON19 dataset (avg. F1-scores of 0.78-0.79), indicating that our approach is on par with standard RGB when training and testing on single-stained IHC data with the same chromogen. In addition, the performance of the unmixed-trained network on the differently-stained CRC dataset (avg. F1-score of 0.74) shows that our method makes IHC-trained algorithms color-agnostic, although it did not outperform the artificial simplex IHC approach (avg. F1-score of 0.82). Lastly, the detection performance of the unmixed-trained network on the PIN and MSKCC datasets (avg. F1-scores of 0.62 and 0.72) demonstrates that our method makes algorithms applicable to mIF imaging, even more so than the artificial bright-field IHC approach (avg. F1-scores of 0.50 and 0.44). In conclusion, we developed an approach for making bright-field IHC deep learning models in digital pathology color-agnostic and applicable to mIF imaging.
The final report is going to be submitted as a conference paper to the International Symposium on Biomedical Imaging (ISBI) 2024 and will hence be added later. The code for this project can be found in this GitHub repository.


