Start date: 22-03-2021
End date: 22-09-2021
Clinical Problem
General practitioners (GPs) work with probabilities of diagnoses to head their diagnostic and therapeutic decisions. To a large extent, this is an implicit process, controlled by prior knowledge and so-called pattern recognition. Less is known about the concreteness and preciseness of the used probabilities. Uncertainty may lead to an overestimation of the probability of a rare disease, and thus lead to overuse of diagnostic facilities, unnecessary costs, and ultimately patient harm. Underestimating probabilities may lead to late diagnoses and detrimental consequences. The process of coming to a diagnosis starts when the patient tells his first complaint. This first utterance of a complaint during the consultation is called reason for encounter (RFE; for example cough, back pain, headache). The RFE itself is related to probabilities of diagnoses.
For common reasons to visit a doctor, context variables influence probabilities of diagnoses. Deepening of our understanding of how diversity, context, multimorbidity and symptoms influence probabilities of final diagnoses will help doctors to work more secure and evidence based. It will lead to the development of a diagnostic support tool to use in everyday practice.
Data
In this project we used data from the research network FaMeNet. More specifically, TransHis was used, which is an Electronic Patient Record (EPR) for primary care. The International Classification of Primary Care (ICPC) is used in the EPR to encode various information such as the RFE, diagnoses, interventions, and referrals. The EPR promotes an Episode of Care structure, where episodes group patient visits that pertain to the same health problem according to the GP.
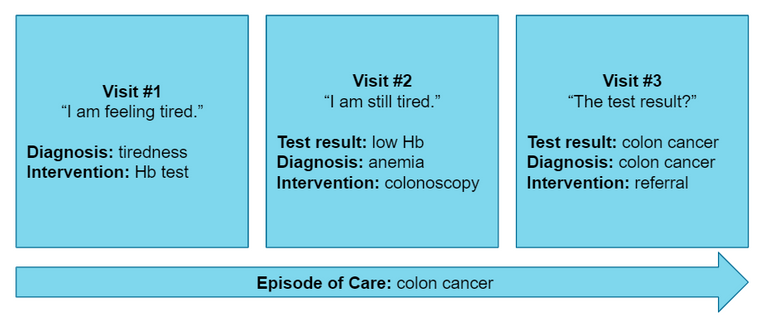
A dataset was created by extracting new episodes (2010-2020) starting with one of the top 15 RFEs for which we wanted to predict the final diagnosis of an episode. The dataset consists of records that describe the first visit of a new episode. The complete dataset contains 143,818 visits from 35,754 patients after preprocessing. Furthermore, historical patient visits (2005-2020) were available in the EPR to be used as contextual information.
Methods
Contextual variables were taken into account by incorporating information from the EPR. A patient's medical history was extracted from the EPR to serve as contextual information. We proposed to represent the patient history as a sequence of episodes, where episodes group related patient visits. A recurrent neural network (RNN) was used to handle the sequential patient history. The RNN encodes the patient history into a latent patient representation, which a classification network uses as a feature vector together with information from the current visit (sex, age, RFE) to predict the probabilities of diagnoses for the current visit.
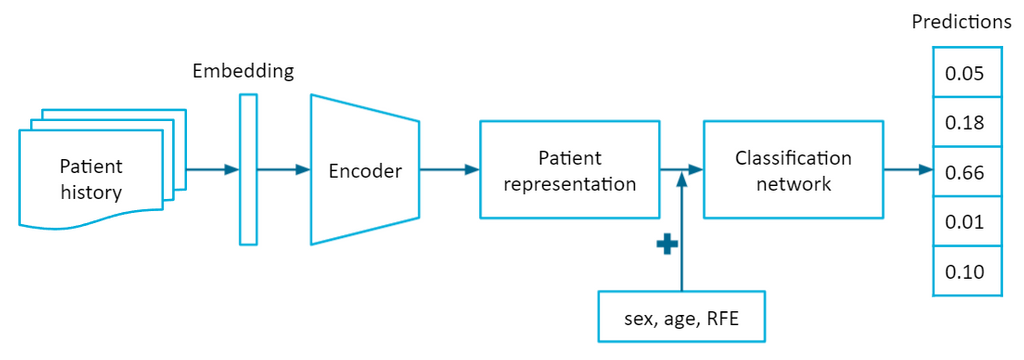
The RNN-based model was compared to a conditional probability baseline for which the conditional probabilities were computed by P(diagnosis | sex, age, RFE) from the data. The conditional probability baseline reflects the current situation in general practice the most in terms of a diagnostic support tool. The other three baselines were logistic regression (LR), XGBoost (XGB), and a multilayer perceptron (MLP), where the patient history is represented by a one-hot vector instead of a sequence.
We further investigated the relevance of time irregularity in the episodic patient history. We considered four conditions with different time assumptions:
- None (MLP): the patient history is represented without retaining the order of episodes and the time between them.
- Weak (LSTM): the patient history is represented as an ordered sequence of episodes, but the time between them is unspecified.
- Moderate (LSTM+∆t): the patient history is represented as an ordered sequence of episodes, and each episode is provided with a time interval feature, which indicates the time since the previous episode.
- Strong (T-LSTM): the patient history is represented as an ordered sequence of episodes, and each episode is provided with a time interval feature, which indicates the time since the previous episode, and the encoder specifically handles irregular time in the sequence.
Condition 1 corresponds to the MLP baseline. Conditions 2 to 4 use the RNN-based model, but the encoder varies. Condition 2 and 3 use an LSTM encoder, whereas condition 4 uses a time-aware LSTM or T-LSTM for short.
Results
| Micro F1 | Macro F1 | Weighted F1 | Acc@3 | Acc@10 | Brier Score | Log Loss | |
|---|---|---|---|---|---|---|---|
| CP | 0.3917 | 0.0496 | 0.2888 | 0.6729 | 0.8761 | 0.7551 | 2.8868 |
| LR | 0.3835 | 0.0605 | 0.3172 | 0.6667 | 0.8678 | 0.7644 | 2.3278 |
| XGB | 0.3979 | 0.0645 | 0.3239 | 0.6809 | 0.8815 | 0.7480 | 2.1897 |
| MLP | 0.3902 | 0.0490 | 0.3020 | 0.6733 | 0.8801 | 0.7530 | 2.1969 |
| LSTM | 0.3940 | 0.0544 | 0.3082 | 0.6795 | 0.8821 | 0.7521 | 2.1790 |
| LSTM+∆t | 0.3974 | 0.0521 | 0.3098 | 0.6798 | 0.8813 | 0.7530 | 2.1833 |
| T-LSTM | 0.3917 | 0.0472 | 0.2880 | 0.6701 | 0.8794 | 0.7548 | 2.1878 |
LSTM performed the best in probabilistic prediction as it achieved the best log loss. XGB performed the best overall in terms of F1-score and accuracy at k. Note that a lower log loss is not necessarily paired with an increase in F1-score or accuracy at k. Accounting for time irregularity in the episodic patient history does not seem to matter much here as LSTM+∆t and T-LSTM either perform comparably to or worse than LSTM. All models performed very poorly on macro F1-score due to the large class imbalance of diagnoses.
Conclusion
We developed a deep learning model that can perform probabilistic prediction of diagnoses for the top 15 most frequent RFEs. Including contextual information from EPRs resulted in improved probabilistic performance and accuracy. Although the increase in performance is rather modest, the superior performance does show that the EPR is beneficial for diagnosis prediction in combination with machine learning and warrants further research. Future work should also focus on how to provide better interpretability for these deep learning models to GPs, and how to properly integrate such models into a clinical decision support system.
The final report can be found here.





