Start date: 22-03-2021
End date: 22-09-2021
Clinical Problem
General practitioners (GPs) work with probabilities of diagnoses to head their diagnostic and therapeutic decisions. To a large extent, this is an implicit process, controlled by prior knowledge and so-called pattern recognition. Less is known about the concreteness and preciseness of the used probabilities. Uncertainty may lead to an overestimation of the probability of a rare disease, and thus lead to overuse of diagnostic facilities, unnecessary costs, and ultimately patient harm. Underestimating probabilities may lead to late diagnoses and detrimental consequences. The process of coming to a diagnosis starts when the patient tells his first complaint. This first utterance of a complaint during the consultation is called reason for encounter (RFE; for example cough, back pain, headache). The RFE itself is related to probabilities of diagnoses.
For common reasons to visit a doctor, context variables influence probabilities of diagnoses. Deepening of our understanding of how diversity, context, multimorbidity and symptoms influence probabilities of final diagnoses will help doctors to work more secure and evidence based. It will lead to the development of a diagnostic support tool to use in everyday practice.
Solution
We aim to analyze how GPs can be supported in early diagnosis through AI support. For all patients with new episodes (2010-2020) we want to define the predictive value of the RFE for diagnoses. More specifically we want to study which data influence predictive values. This knowledge is crucial to build ICT tools to support the GP.
The AI challenge is to use machine learning (eg Bayesian network approach and other methods) to calculate probabilities of diagnoses based on the reason for encounter, modified for other personal and context variables, based on the data in our database. For 15 common reasons to visit the GP, we want to develop an algorithm that is able to show a physician realtime for any unique patient the probabilities of diagnoses.
Data
We will use data from the research network FaMeNet (www.famenet.nl) covering over 300.000 patient years, and over 1 million patient contacts. Structured data on contextual factors are available for more than 50% of the adult population. Contextual factors include chronic comorbidity, sex, age, ethnicity, educational level, and all symptoms and diagnoses in the two years preceding the diagnosis.
The data are stored in a data warehouse at the department of Primary and Community care of the Radboud University Nijmegen Medical Center (Radboud Technology Center Health Data).
For the purpose of this research we will only consider the top 15 most common RFEs. That is, we only consider patients that have at least 1 episode starting with one of the top 15 RFEs. Each data point consists of the following patient information: RFE, age, sex, start date and diagnosis. We augment this data by modelling the patient history up to the current RFE and add it as a feature to the input. By doing this we are trying to simulate the way in which general practitioners use a patient's history, combined with current demographics to find the correct diagnosis. Important to note is that it is possible for a patient to appear multiple times in the data, at different moments in time. In those cases there is some overlap in patient history in the data, but the demographics will differ (such as RFE and age).
Approach
To be able to model the patient history, we will use the BERT architecture which is based on the Transformer architecture which is widely adopted in NLP applications. This network is already used once in the same domain for a similar problem, where it was renamed BEHRT: Transformer for Electronic Health Records. The method is visualized in the image below.
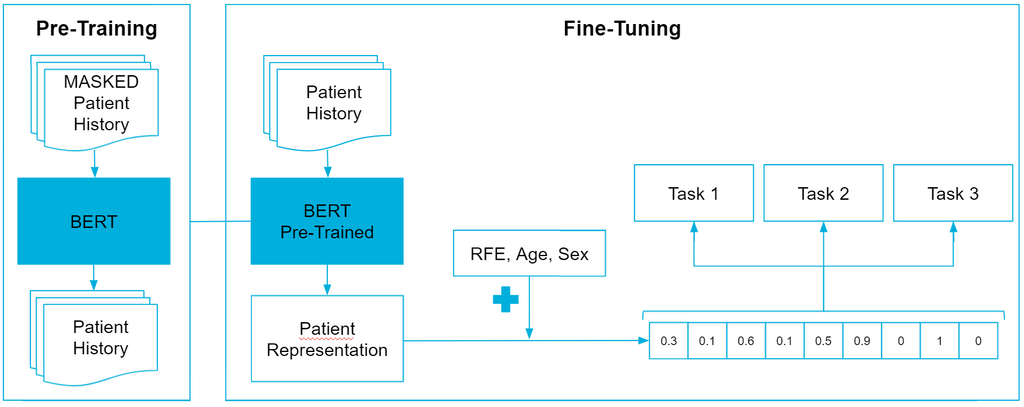
First off, we need to pre-train BEHRT, which is done by applying Masked Language Modeling (MLM). This approach is pretty straightforward, in that we replace a percentage of the input tokens with '
The next step is to fine-tune pre-trained BEHRT. More specifically, we want to experiment with Mult-Task Learning (MTL) to see if we can achieve better performance than without MTL. Inspired by human learning abilities, the MTL learning paradigm aims to jointly learn multiple related tasks so that the knowledge contained in one task can be leveraged by other tasks, hoping to improve generalization performance of all tasks at hand. In a sense MTL can be seen as a form of data augmentation, where the labeled data in all tasks is aggregated. With more data, MTL should be able to learn more robust representations for all tasks, leading to better knowledge sharing among tasks, better performance of each task and a reduced risk of overfitting. The first auxiliary task we selected is the prediction of interventions. Interventions are actions a general practitioner can perform during a contact. This can range from a physical examination to prescribing a recipe or making an appointment for taking an x-ray. The second auxiliary task we selected is predicting whether or not a patient returns for the same health problem on another day.
Results
| Micro F1 | Macro F1 | Weighted F1 | Acc@5 | Acc@10 | Cross-entropy Loss | Brier Score | |
|---|---|---|---|---|---|---|---|
| CPC | 0.3821 | 0.04285 | 0.2727 | 0.7711 | 0.8784 | 2.548 | 0.7598 |
| LR | 0.3779 | 0.05882 | 0.3156 | 0.7632 | 0.8702 | 2.338 | 0.7734 |
| XGBoost | 0.3951 | 0.05816 | 0.3147 | 0.7832 | 0.8815 | 2.332 | 0.7581 |
| MLP | 0.3821 | 0.05801 | 0.3158 | 0.7707 | 0.8801 | 2.241 | 0.7696 |
| BEHRT | 0.3930 | 0.05779 | 0.3203 | 0.7829 | 0.8846 | 2.178 | 0.7522 |
| Multi-BEHRT | 0.3944 | 0.05939 | 0.3197 | 0.7823 | 0.8847 | 2.170 | 0.7518 |
The scores for all models on the metrics are quite close to each other. Specifically, when compared to the simple CPC, we see that on most metrics the absolute differences in performance are not that large. Additionally, when looking at the overall low F1-macro scores, we can conclude that all models have difficulty in predicting more rare diagnoses. This can be attributed to the large class imbalance in the target diagnoses.
In terms of classification accuracy, our model achieves the overall highest F1-macro score. This indicates that our model outperforms all baselines in terms of predicting more rare diagnoses. Compared with the second best performance on both F1-micro and F1-weighted, we are able to conclude that our model is better than most baselines in both predicting common diagnoses, as well as more rare diagnoses.
In terms of probabilistic predictions, our model outperforms all baselines on both the cross-entropy loss as well as the Brier score. This means that the predicted probabilities are closest to the targets. Combined with the best performance on the top-10 accuracy, this indicates that in terms of performance, our model would be best suited for a clinical support system that predicts the top-10 diagnoses along with their probabilities.









