Clinical Problem
The primary bottleneck for interventional MRI is accurately positioning instruments in the correct position relative to the lesion. The instruments, operated by hand or robots, move unpredictably and currently require a careful and iterative process of move - scan - move - scan until the position is satisfactory. Conversely, the lesion moves much less and can generally be assumed to stay within a region.
Guiding a percutaneous needle towards the region of interest in a prostate biopsy may take several minutes per biopsy. Cryoablation interventions, for example, use several needles and may take hours to position correctly. MRI-guided interventions can be made faster and more accurate by developing solutions allowing for real-time acquisitions, greatly improving current time-consuming processes.
Method
Based on previous work showing dynamic deep-learning algorithms capable of reconstructing highly undersampled acquisitions, we modified a BCRNN-MRI algorithm to reconstruct the sagittal and transverse images of a pelvis containing a biopsy needle in a dynamic fashion. An ablation study was performed to show that the BCRNN model is able to exploit the temporal information in interventional MR images. The BCRNN model is trained on images of undersampling factors 8,16,25 and 32 and on two network sizes. Next to the BCRNN model we integrated a segmentation model to quanitify how well the needle guide is visible in the reconstructed images. In this project not much effort is put in creating the most optimized models.
Data
The dataset consists of 4338 sagittal and 4315 transverse image volumes of shape 256x256x5. For training/validation, 500 sagittal images were used. 200 images were used in the testset. The dataset contains biopsy images made right after the biopsy was performed. No needle movement is visible in the images. To simulate needle movement, we used the 5-slice volumes as if they were a time sequence, where the needle moves in the z-axis.
Experiment - Ablation Study
The results for the Simulated Temporal Neural Network (STNN and STNN-L(arge)) and Non-Temporal Ablated Neural Network (NANN and NANN-L) are shown below.
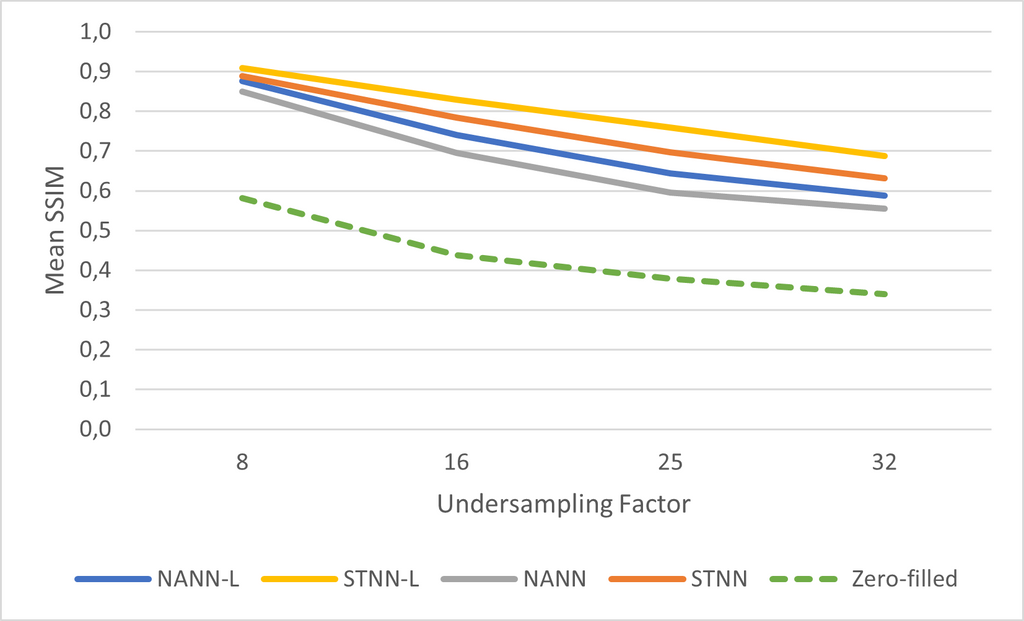 The results show that the network is able to exploit the temporal information from the input images. In all experiments the STNN outperformed the NANN network by a significant margin (p<0.01). The dotted green line indicates the zero-filled SSIM where no reconstruction is done. The performance of the larger network is better than the smaller network. The small network has an average inference speed of 20ms compared to 135ms for the large network.
The results show that the network is able to exploit the temporal information from the input images. In all experiments the STNN outperformed the NANN network by a significant margin (p<0.01). The dotted green line indicates the zero-filled SSIM where no reconstruction is done. The performance of the larger network is better than the smaller network. The small network has an average inference speed of 20ms compared to 135ms for the large network.
Experiment - Segmentation
An nnU-Net was trained on 200 manually annotated sagittal image volumes, with a training DICE of 0.95. For all models trained in the previous experiment, segmentations were made on 190 reconstructed images. Results can be seen below.
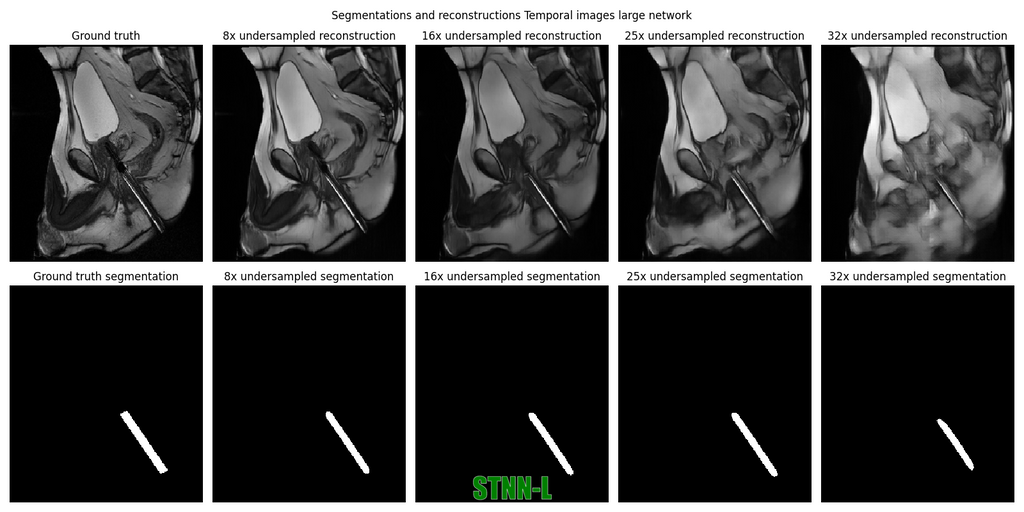
 The segmentation model is able to segment the needle from the reconstructions made by the STNN-L network for undersampling of up to 25x. For the Non-Temporal networks, the performance is good at 8x undersampling, but gets worse when undersampling increases.
Example reconstructions and segmentations can be seen here:
The segmentation model is able to segment the needle from the reconstructions made by the STNN-L network for undersampling of up to 25x. For the Non-Temporal networks, the performance is good at 8x undersampling, but gets worse when undersampling increases.
Example reconstructions and segmentations can be seen here:
Conclusion
In this project we have shown that the BCRNN is a suited model to exploit temporal information in interventional MR images. The model is able to reconstruct images of up to 25x undersampling while maintaining a DICE score of 0.79 for needle guide segmentation. With some optimalizations in inference speed, this model can be used to provide real-time imaging during biopsies.
Thesis can be found in the Radboud Master Archive
The code for this project can be found on GitHub

