Start date: 28-11-2022
End date: 28-09-2022
Clinical Problem
Acute myocardial infarction (MI), also known as a “heart attack”, is a life threatening condition that occurs when blood supply to the heart is decreased or stopped, usually as a result of narrowing / blockage of a coronary artery due to atherosclerotic disease. In patients with MI, coronary angiography (CAG) is usually performed to localize the occluded artery, known as the culprit lesion, and to treat this artery by placing a stent (Dotter procedure). In some cases, more than one lesion is found during CAG. As of yet, it remains largely unknown whether it is beneficial to treat these non-culprit lesions in patients with so-called multi-vessel disease. To date, we cannot accurately predict which atherosclerotic plaques are vulnerable and have a high risk of causing an MI in the future, and which plaques are relatively safe.
Optical coherence tomography (OCT) is a relatively new imaging modality within interventional cardiology and cardiovascular research. OCT uses near-infrared light and is used to provide a high-resolution intravascular image of the coronary artery, allowing for assessment of plaque morphology and characteristics. Previous research has demonstrated its potential on several key aspects in the management of patients with atherosclerotic coronary artery disease, leading to beneficial patient outcome. Hence, OCT is a potential game changes in the field of interventional cardiology, as it may serve as a future tool to distinguish ‘high risk’ from ‘safe’ plaques in patients with acute MI and multi vessel disease.
Currently, however, the use of OCT in daily clinical practice is limited due to a lack of training for assessment and analysis of OCT-acquired pullbacks. In the current project, we aim at easing the use of OCT by means of artificial intelligence (AI), thereby increasing the availability of AI-based OCT algorithms for clinical decision making, and eventually improving patients’ health.
Solution
A semantic segmentation algorithm based on the no-new U-Net (nnU-Net) architecture was developed that segments 12 different targets (lumen, guidewire, intima, lipid, calcium, media, catheter, sidebranch, red and white thrombus, dissection and plaque rupture) in intracoronary OCT scans. A total of 4 models were trained: one of them was trained on 2D OCT frames, and the other three were trained on pseudo 3D input, in which k frames before and after the frame with the ground truth annotation are also included as input. Thus, we trained these three models using k = 1, k = 2 and k = 3 frames before and after. As preprocessing steps, a circular mask and resizing interpolator are employed to create a curated dataset suitable for training. The figure below shows the preprocessing and training framework, considering the 2D and pseudo 3D with k = 3 cases
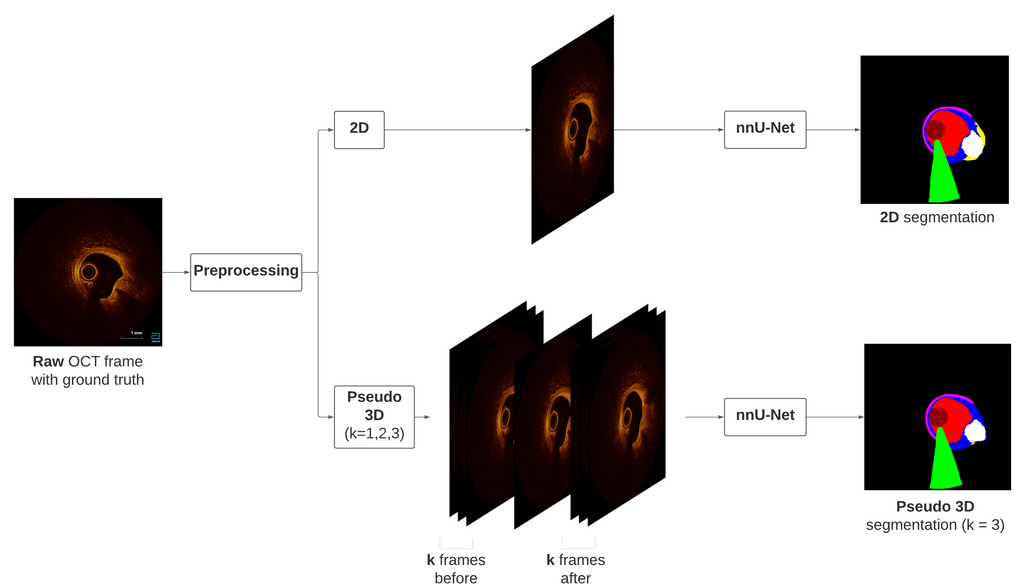
Preprocessing and training frameworks, for the 2D case and the k = 3 case for the pseudo 3D model.
After the model training, a post-processing framework based on automatic lipid and calcium measurements is designed, based on the predicted segmentations. This automated analysis measures the Fibrous Cap Thickness (FCT) and lipid arc for lipid, assesing for Thin-Cap Fibroatheroma (TCFA) detection, and the calcium arc, thickness and depth for calcium. A threshold for lipid and calcium size is estimated by computing the ROC curves and finding the mimumum nº of pixels that a lipid or calcium region must contain in order to consider it as such. A final analysis addresses for the "black-box" problem that many DL models suffer. This is done by retrieving the features maps after each convolutional layer for Explainable AI (XAI). For uncertainty estimation, the reliability curves and the total Expected Calibration Error (ECE), including the ECE for lipid and calcium, for the test set are obtained. Further uncertainty analysis on the final probability maps are performed in order to validate these maps as a measure for uncertainty, focusing into lipid and calcium regions. The entropy per pixel is also obtained in order to perform correlation analysis between the entropy on lipid and calcium regions with the DICE score. The figure below shows this post-processing framework for a predicted segmentation.
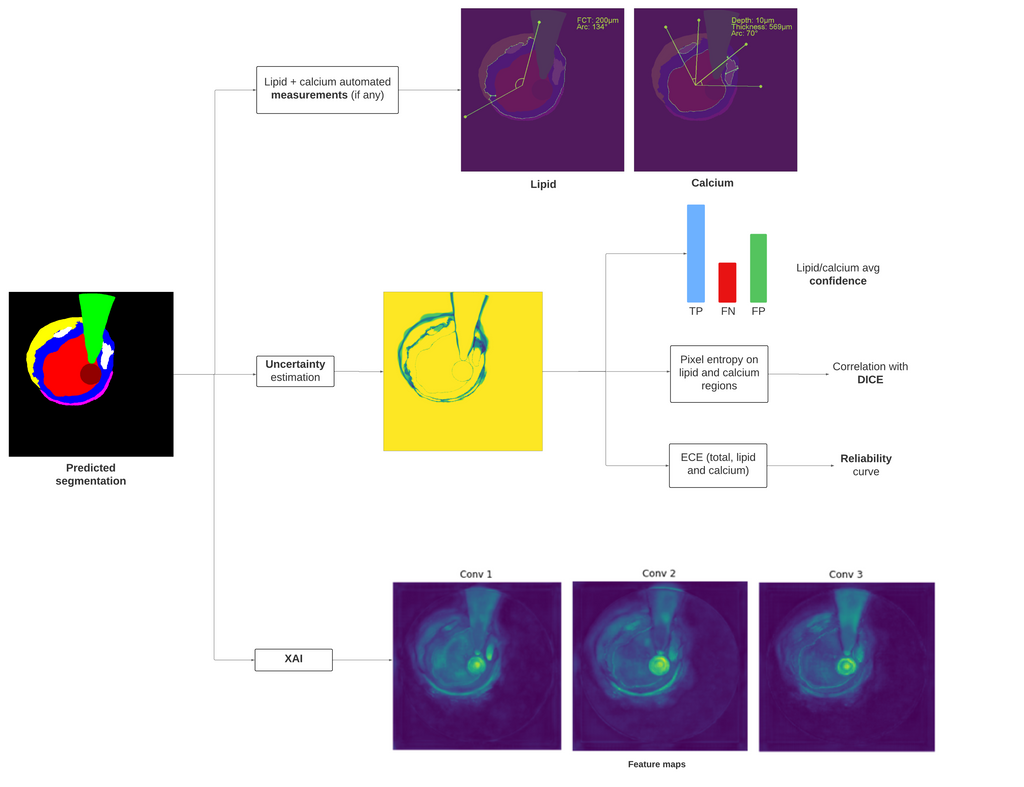
Post-processing framework, from the automated measurements on lipid calcium, to XAI and uncertainty estimation analysis.
Data
We will use OCT data from the PECTUS-obs study (https://pubmed.ncbi.nlm.nih.gov/34233996/) to develop and internally validate the algorithm. In total, 2028 OCT frames from this dataset were manually segmented by an expert annotator. A random 9:1 train/test split was performed, obtaining 1810 frames for training and 218 for testing.
Conclusion
In this project, a semantic segmentation model based on the nnU-Net was developed to segment intracoronary OCT scans. By leveraging contiguous frames to the ground truth, a pseudo 3D was also developed, using either 1, 2 or 3 frames before and after the frame with ground truth. While the 2D and pseudo 3D approach seem very similar, it was concluded that the pseudo 3D model with k = 3 frames before and after provided the overall best results. The post-processing algorithm for lipid and calcium automated assesement can be further used to peform fast and accurate measurements, altough further improvements need to be done in order to detect TCFAs in a reliable way. The model transparency could be further analysed and correlated with the model's output, such as by using other techniques like Grad-CAM. Finally, the model showed great reliability in its confidence values, meaning that the confidence maps could potentially be used in clinical practice as an uncertainty map and see which regions of the OCT frame are less confident. The model could be improved by using a 3D network, and for this more annotations should be included, plus including more cases of rarer structures in order to improve the model's capabilities to detect these regions (white thrombus, dissection or plaque rupture). Other improvements would include to tune the train set annotations and include other common regions such as layered plaque, include OCT scans from different scanners ot annotators, or train the algorithm on the original grayscale version of the OCT scans.
You can try the pseudo 3D (k = 3) algorithm on Grand Challenge: Try out the algorithm
The code for this project can be found in the following Github repository.





