Start date: 08-02-2021
End date: 06-08-2021
Clinical problem
This project aims to improve outcomes in the 30,000 patients per year in the Netherlands that develop chronic pain after abdominal surgery. This pain is often caused by adhesions, tough bands of tissue that form between structures and organs in the abdomen. Medical consumption in these patients is high, 70% is under treatment of a gastroenterologist and one in three is permanently taking pain suppressing medication.
Currently, accurate diagnosis of adhesions requires invasive tools such as diagnostic laparoscopy owing to a lack of effective noninvasive tests. These methods are controversial because they can lead to the formation of new adhesions. This is especially undesirable for the patients whose complaints are not due to adhesions. Non-invasively adhesions can be diagnosed with either ultrasound or cine-MRI. Cine-MRI is more powerful than ultrasound because it can detect adhesions in the entire abdomen, whilst ultrasound can detect only adhesions attached to the front abdominal wall due to its limited depth penetration. Cine-MRI is a type of MRI in which a set of consecutive images of the area of interest is acquired at a fixed time interval. The captured images can be merged into a video in which the movements of a certain tissue over time are visible. This imaging modality can be used to visualise the abdominal motion during respiration and by interpreting the recorded motion patterns, radiologists can diagnose adhesions. The gif below shows the data and annotations available in the project. Adhesions are annotated with bounding boxes by an experienced radiologist.
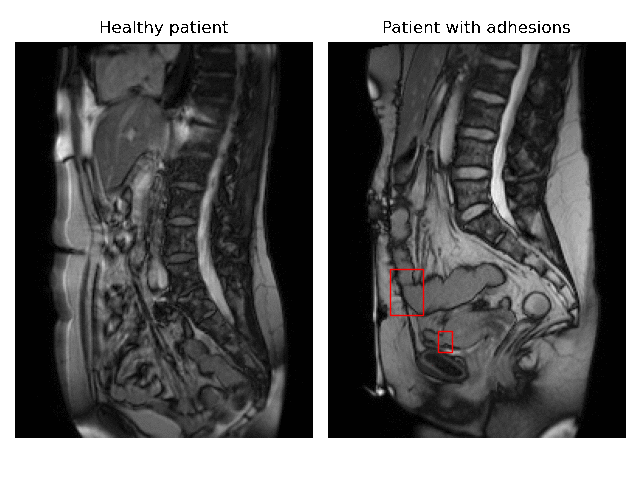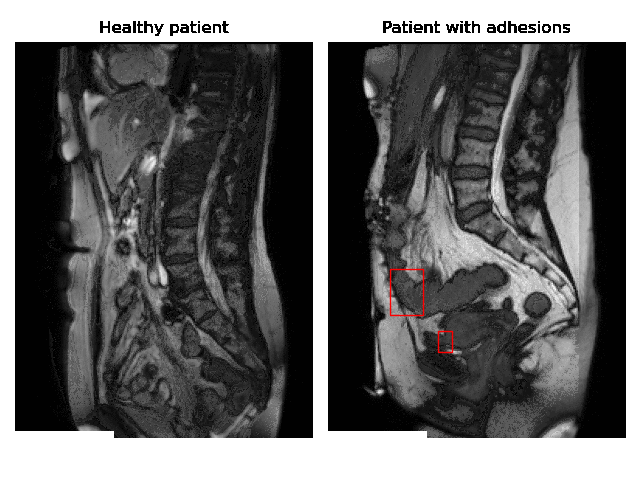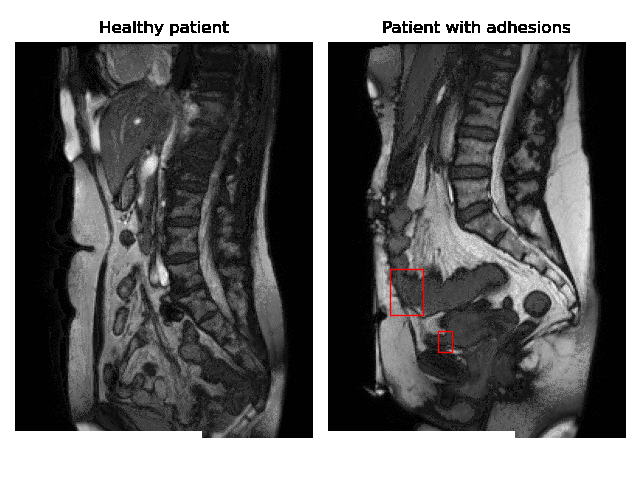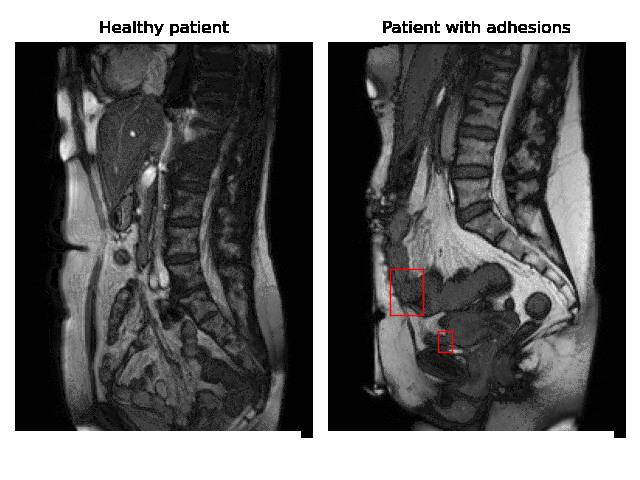
Cine-MRI is used in the Radboudumc to make treatment decisions for patients with chronic abdominal pain. It helps doctors decide whether a patient should be treated conservatively with medication, or more progressively with surgery. Cine MRI is not widely used in other hospitals yet, because radiological reading is time-consuming and expertise-dependent.
Automatic algorithms for adhesion detection can reduce the learning curve for new readers and reduce variability among radiologists. By helping radiologists interpret the images, the algorithm may reduce the number of false positives and negatives. False positives can result in unnecessary surgery, whereas false negatives can block surgery for patients who may actually be helped by it.
Solution
The objective of the project is to design a fully automated CAD method for adhesion detection. As a starting point, we take a semi-automated method proposed by David Randall in his PhD thesis [1]. The method exploits the discontinuity of the abdominal motion during respiration. In healthy subjects, abdominal contents slide smoothly against the surroundings of the abdominal cavity (abdominal wall, back muscles, etc.). Simultaneously, the abdominal wall exhibits a different, anteroposterior mode of motion. This process is called visceral slide. The scheme of this motion pattern is given below.
Reduction in visceral slide is a clinical criterion of underlying adhesions. The key idea of the method is that it is possible to quantify the degree of visceral slide captured on a cine-MRI scan. To do this, the method needs a segmentation map of the abdominal cavity, to perform masked image registration. The output of masked image registration is a deformation field that describes the amount and direction of movement of the abdomen and its contents. Then, the deformation field and abdominal cavity segmentation can be used to quantify visceral slide along the abdominal cavity contour.
One of the limitations of the method is the lack of segmentation automation. For the first cine-MRI frame a segmentation map has to be drawn by a medical specialist and then it is transferred to the subsequent frames. But manual correction is still required for each frame. Besides, the method assumes the manual interpretation of the computed visceral slide by a radiologist. We aimed at full automation of this method using deep learning-based segmentation of the abdominal cavity and designing an algorithm that predicts adhesions based on the low visceral slide value.
Data
The entire dataset available for the project contains 563 cine-MRI scans for 526 patients. Each scan is provided with a report containing a radiologist’s diagnosis based on the assessment of a cine-MRI scan. From the conclusions in the report, binary study-level annotations were extracted. This dataset was used to sample training and test sets balanced by patient adhesion status as well as a healthy control group to calculate visceral slide statistics. To acquire bounding box annotations of adhesions, two reader studies were organised separately for training and test sets. The resulting training set consists of 59 cine-MRI scans with 83 adhesions annotations and 59 negative scans. The held-out test set contains 38 scans with 56 adhesions annotations and 47 negative scans. Finally, 36 negative scans were sampled for the healthy control group.
Method
We propose the first fully-automated multi-stage CAD method for adhesion detection on cine-MRI designed based on Randall's semi-automated method. In addition, multiple normalization options that account for motion differences between patients and in different areas of abdominal cavity contour were carefully designed and integrated. This enables the same interpretation of a particular visceral slide value regardless of the amplitude of motion that occurs on a cine-MRI slice and the position at which it is observed. Also, two different approaches to visceral slide quantification were explored. In the first one, only the two most dissimilar frames of a cine-MRI scan are used to sharpen the differences between regions of lower and higher visceral slide and the second method exploits all time points in a cine-MRI slice to obtain a full picture of motion.
The full scheme of the method is visualised below:
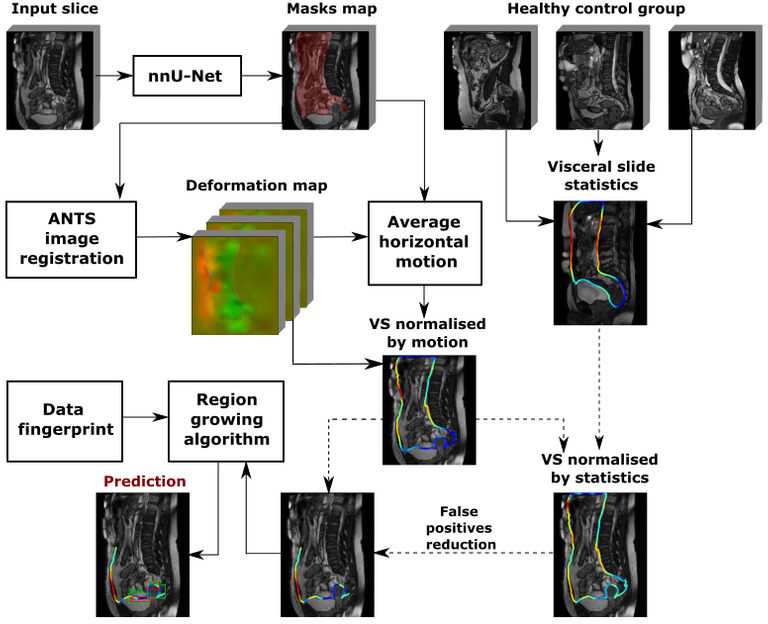
The main components of the methods are:
Abdominal cavity segmentation
Implemented using nnU-Net [2], a state of the art model for medical image segmentation.
Visceral slide computation
A predicted segmentation mask of the abdominal cavity is used to obtain a masked deformation field and compute visceral slide along the abdominal cavity contour based on the difference of deformation inside and outside of the abdominal cavity. We used the ANTS toolkit [3] to obtain a deformation field. The first method to quantify visceral slide uses only the two most dissimilar frames, that correspond to the opposite phases of the respiratory cycle, to approximate the visceral slide that occurs on a scan. The picture below visualises the algorithm:
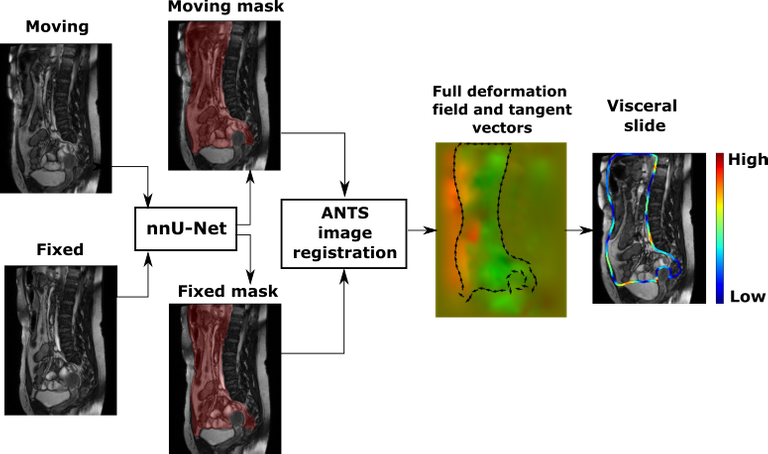
Another method we explored extracts cumulative visceral slide from a cine-MRI scan by adding visceral slide maps computed between each subsequent pair of frames with a specially designed summation procedure. We assumed that the latter method captures the full motion recorded in a cine-MRI scan more accurately.
Visceral slide normalisation
Despite receiving the same instructions during a cine-MRI scan acquisition, different patients move with different amplitude. Some patients exhibit high amplitude motion, whereas others show little motion. We mitigate motion differences between patients by normalising the computed visceral slide by the average motion that occurs on the scan.
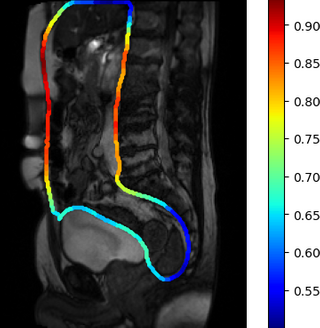
Similar normalisation is necessary to account for normal motion amplitude in different parts of the abdomen. For instance, generally, the motion in the pelvis is noticeably lower than motion along the abdominal cavity walls, which affect the computed visceral slide values in these areas. Suitable normalisation was performed with the visceral slide statistics by position on the abdominal cavity contour in the healthy control group. Two normalisation options were tried: division of visceral slide by expectation and standardisation. The visceral slide expectation that we obtained for different areas of the abdominal cavity is visualised in the picture below:
Adhesion detection with region growing algorithm
Finally, the adhesions are predicted based on the normalized visceral slide map with a custom region growing algorithm. The first step of the algorithm is the false positive reduction based on domain knowledge. This is done by filtering out the parts of the abdominal cavity contour where adhesions cannot form. Hence, only the bottom half of the front abdominal wall and pelvis area are considered during prediction. In each iteration, the algorithm starts searching for adhesion from the minimum value of the visceral slide. Then, a bounding box is grown until one of the stop criteria has been reached. The process is repeated until no points are left in the abdominal cavity contour. Each prediction is assigned a confidence score that can be used to filter prediction by thresholding. Three ways to output confidence have been tried: minimum value of visceral slide inside a bounding box, mean visceral slide value inside a bounding box and prediction of the logistic regression trained with 5-fold CV on the set of features extracted from bounding boxes.
An example of the algorithm output is given below.
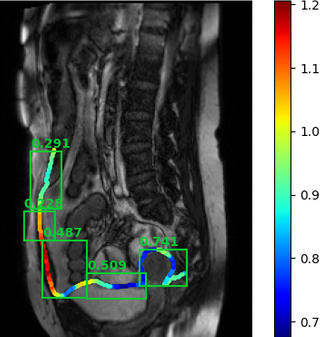
Results
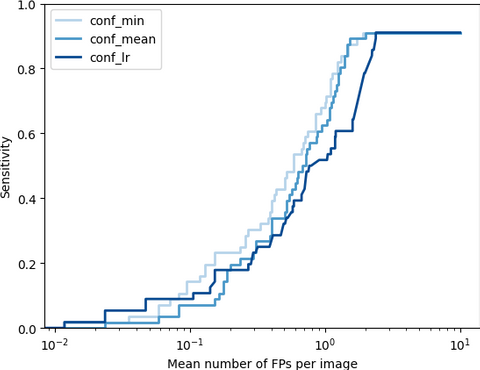
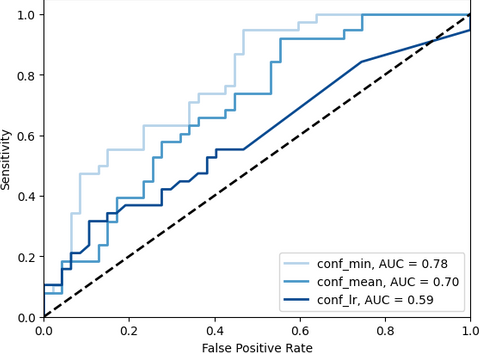
To evaluate the adhesion detection task we used average precision (AP) and FROC analysis. Additionally, the ability of the method to distinguish healthy patients and patients with adhesions is accessed with slice-level ROC. We found that usage of cumulative visceral slide normalised with division by control group expectation yields the best performance. On the held-out test set, the method achieves detection sensitivity of 0.7 and 0.91 at 1 and 1.89 false positives per slice along with 0.78 slice-level AUC, which indicates the promising potential of the core idea of the method.
The FROCs on the test set for cumulative visceral slide normalised with division by control group expectation. The methods are different with respect to the way to output confidence: min - minimum value of visceral slide inside a bounding box, mean - mean visceral slide value, lr - predicted with logistic regression.
The corresponding slice-level ROCs:
Since the performance of the method is insufficient to be proposed for usage in clinical practice, we decided to only deliver a visceral slide map as a tool to assist in adhesion diagnosis. The visceral slide map is visualised over one of the cine-MRI scan's frames.
The algorithm is published on the Grand Challenge platform. Visualisation of the output in a viewer is not supported now. To view the result, please click on the image.mha link in the "Result" column, go to "View Result Details" and download output files. Visualised visceral slide map is saved as a .tif file and can be opened with an image viewer.
The master's thesis that describes the metod and results in details can be found here.
The source code of the project including the complete method is published on GitHub.
[1] Randall, David. "Towards a non-invasive diagnostic aid for abdominal adhesions using dynamic MRI and image processing." PhD diss., University of Sheffield, 2017.
[2] Isensee, F., Jaeger, P.F., Kohl, S.A., Petersen, J. and Maier-Hein, K.H., 2021. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature Methods, 18(2), pp.203-211.
[3]Avants, B.B., Tustison, N.J., Song, G. and Gee, J.C., 2009. Ants: Open-source tools for normalization and neuroanatomy. HeanetIe, 10, pp.1-11.


