Reimbursement
This is an AI for Health MSc project. Students are eligible to receive a monthly reimbursement of €500,- for a period of six months. For more information please read the requirements.
Clinical Problem
Millions of women undergo breast biopsies (i.e., tissue sampling) each year to determine whether a suspicious lesion found on a mammogram (e.g., calcifications, a mass or density) or on physical examination represents breast cancer (BC). Although 75% of breast biopsies show benign breast disease (BBD), which is reassuring, women with BBD experience a 1.5- to four-fold increased risk of future development of BC compared with women not diagnosed with BBD. To better identify women that would benefit from intensified monitoring, improved prediction of BC risk is urgently needed.
Project goal
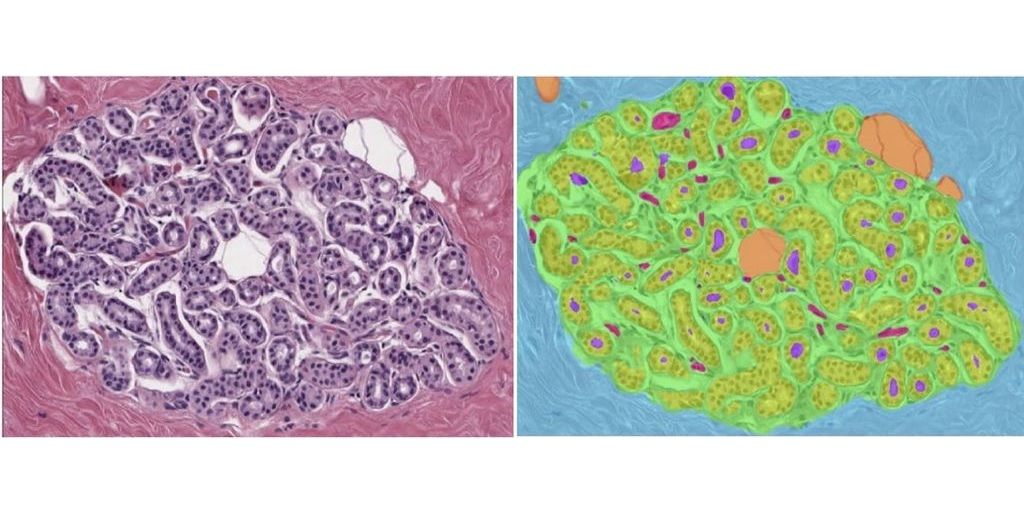
Convolutional neural networks to characterize tissue composition of biopsies and assess the morphology of normal breast lobules (which are the source of BBD) have been developed and reported in several publications (PMC9275035; PMC8770616; PMID: 35503494). The goals of this project include: - characterization of morphologic features of the BBD biopsies - establishing approaches for predicting BC risk.
Data
Mayo Clinic (Rochester, US) has established a cohort of 5,499 women biopsied for BBD between 2002-2013, including 360 who developed an incident DCIS or invasive BC. Microscopic slides of all biopsies have been thoroughly reviewed and analyzed and digital images of a subset have been incorporated into a case-cohort, which includes all biopsies preceding BC and a random subset of biopsies (~900 total women).
Results
Thesis, source code, documentation; the algorithm will be made publically available as a Docker container on https://grand-challenge.org/.
Embedding
The student will be supervised by a research member of the Diagnostic Image Analysis Group and Computational Pathology Group whose research is dedicated to analyses of histopathological slides with deep learning techniques. We have a strong collaboration with pathology experts in the field of cancer grading. The student will have access to a large GPU cluster.
Requirements
- Students with a major in computer science, biomedical engineering, artificial intelligence, physics, or a related area in the final stage of master level studies are invited to apply.
- Affinity with programming in Python
- Interest in deep learning and medical image analysis
Information
- Project duration: 6 months
- Location: Radboud University Nijmegen Medical Center
- For more information please contact Witali Aswolinskiy or Jeroen van der Laak


